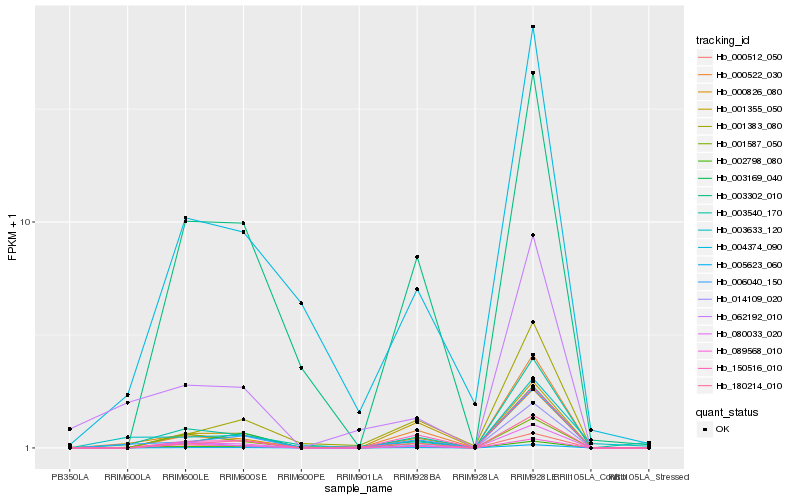
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000826_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_006040_150 |
0.1126145005 |
- |
- |
- |
| 3 |
Hb_080033_020 |
0.1615539149 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 4 |
Hb_005623_060 |
0.1617739702 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000512_050 |
0.163751556 |
- |
- |
hypothetical protein VITISV_005640 [Vitis vinifera] |
| 6 |
Hb_003540_170 |
0.168759646 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_150516_010 |
0.1750512708 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_062192_010 |
0.179405452 |
- |
- |
- |
| 9 |
Hb_003302_010 |
0.1832906631 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 10 |
Hb_001355_050 |
0.18911428 |
- |
- |
- |
| 11 |
Hb_000522_030 |
0.1931930772 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 12 |
Hb_004374_090 |
0.1996560516 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 13 |
Hb_180214_010 |
0.2001332694 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 14 |
Hb_089568_010 |
0.2003765386 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 15 |
Hb_001587_050 |
0.2004285754 |
- |
- |
- |
| 16 |
Hb_003169_040 |
0.2025902073 |
- |
- |
- |
| 17 |
Hb_014109_020 |
0.2068940693 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 18 |
Hb_003633_120 |
0.2076643924 |
- |
- |
PREDICTED: uncharacterized protein LOC104880778 [Vitis vinifera] |
| 19 |
Hb_002798_080 |
0.2132579252 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 20 |
Hb_001383_080 |
0.2174388884 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |