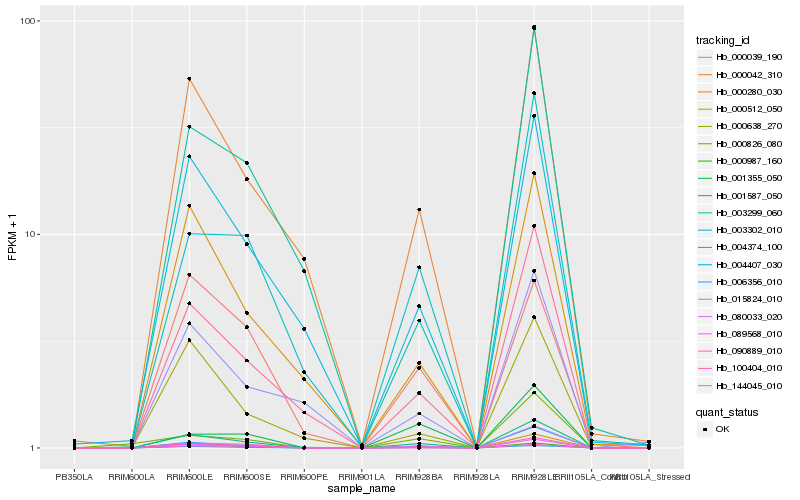
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001587_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_089568_010 |
0.0552597851 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 3 |
Hb_006356_010 |
0.0613182152 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 4 |
Hb_144045_010 |
0.131675642 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 5 |
Hb_080033_020 |
0.1341851675 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 6 |
Hb_000042_310 |
0.1491980718 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 7 |
Hb_090889_010 |
0.1514504797 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 8 |
Hb_003302_010 |
0.1605167905 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 9 |
Hb_004374_100 |
0.1632398425 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 10 |
Hb_000280_030 |
0.1670793622 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 11 |
Hb_000638_270 |
0.1712475701 |
- |
- |
- |
| 12 |
Hb_000987_160 |
0.1809108323 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 13 |
Hb_003299_060 |
0.1906947537 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_000512_050 |
0.1909943495 |
- |
- |
hypothetical protein VITISV_005640 [Vitis vinifera] |
| 15 |
Hb_015824_010 |
0.1937259819 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 16 |
Hb_000826_080 |
0.2004285754 |
- |
- |
- |
| 17 |
Hb_001355_050 |
0.2013180614 |
- |
- |
- |
| 18 |
Hb_100404_010 |
0.2022145897 |
- |
- |
Cysteine-rich receptor-like protein kinase 29 [Glycine soja] |
| 19 |
Hb_004407_030 |
0.2061717405 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 20 |
Hb_000039_190 |
0.2092046075 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |