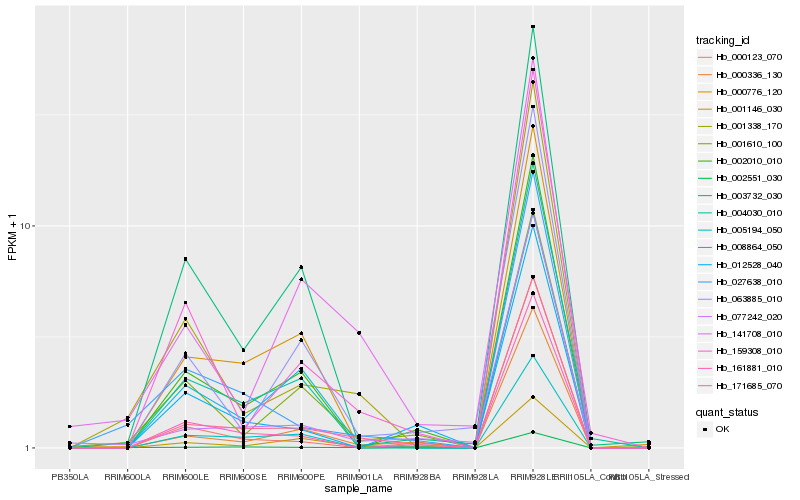
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171685_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000336_130 |
0.0930951066 |
- |
- |
Triacylglycerol lipase 2 precursor, putative [Ricinus communis] |
| 3 |
Hb_161881_010 |
0.1070977844 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_002010_010 |
0.1086279801 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 5 |
Hb_002551_030 |
0.1177760544 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 6 |
Hb_027638_010 |
0.1248783075 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 7 |
Hb_003732_030 |
0.126244998 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 8 |
Hb_000776_120 |
0.1262685422 |
- |
- |
PREDICTED: cytochrome P450 78A5 [Jatropha curcas] |
| 9 |
Hb_004030_010 |
0.126292691 |
- |
- |
hypothetical protein CICLE_v10013050mg [Citrus clementina] |
| 10 |
Hb_005194_050 |
0.131073974 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 11 |
Hb_159308_010 |
0.1314872565 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 12 |
Hb_001338_170 |
0.132342167 |
- |
- |
NADH dehydrogenase subunit 4 [Hevea brasiliensis] |
| 13 |
Hb_001610_100 |
0.1339434129 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_077242_020 |
0.13403569 |
- |
- |
ribosomal protein S4 (mitochondrion) [Ricinus communis] |
| 15 |
Hb_000123_070 |
0.1372237238 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 16 |
Hb_012528_040 |
0.1374732535 |
- |
- |
- |
| 17 |
Hb_063885_010 |
0.1388919857 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_008864_050 |
0.1433864755 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_001146_030 |
0.1435700877 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_141708_010 |
0.1437106838 |
- |
- |
hypothetical protein EUTSA_v10011731mg [Eutrema salsugineum] |