| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
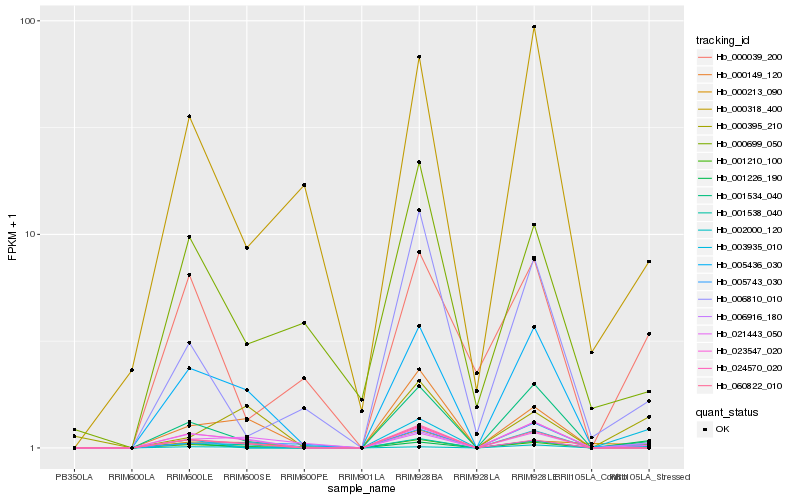
Hb_001210_100 |
0.0 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 2 |
Hb_006916_180 |
0.1957272545 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 3 |
Hb_001534_040 |
0.2090551716 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 4 |
Hb_003935_010 |
0.2325000387 |
- |
- |
- |
| 5 |
Hb_002000_120 |
0.2459239074 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_005743_030 |
0.2534079033 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 7 |
Hb_005436_030 |
0.2558468451 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74E1-like [Jatropha curcas] |
| 8 |
Hb_060822_010 |
0.2568257798 |
- |
- |
- |
| 9 |
Hb_000213_090 |
0.2602266191 |
- |
- |
hypothetical protein OsI_39056 [Oryza sativa Indica Group] |
| 10 |
Hb_001538_040 |
0.265584672 |
- |
- |
PREDICTED: uncharacterized protein LOC104228304 [Nicotiana sylvestris] |
| 11 |
Hb_000039_200 |
0.2702662762 |
- |
- |
hypothetical protein JCGZ_21346 [Jatropha curcas] |
| 12 |
Hb_001226_190 |
0.2764697349 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 13 |
Hb_006810_010 |
0.2777840745 |
- |
- |
hypothetical protein B456_012G018200 [Gossypium raimondii] |
| 14 |
Hb_024570_020 |
0.2805956015 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 15 |
Hb_000395_210 |
0.2849383159 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 16 |
Hb_023547_020 |
0.2862608531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_021443_050 |
0.288189918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000149_120 |
0.2919641634 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000699_050 |
0.2976259387 |
- |
- |
- |
| 20 |
Hb_000318_400 |
0.3007183145 |
- |
- |
conserved hypothetical protein [Ricinus communis] |