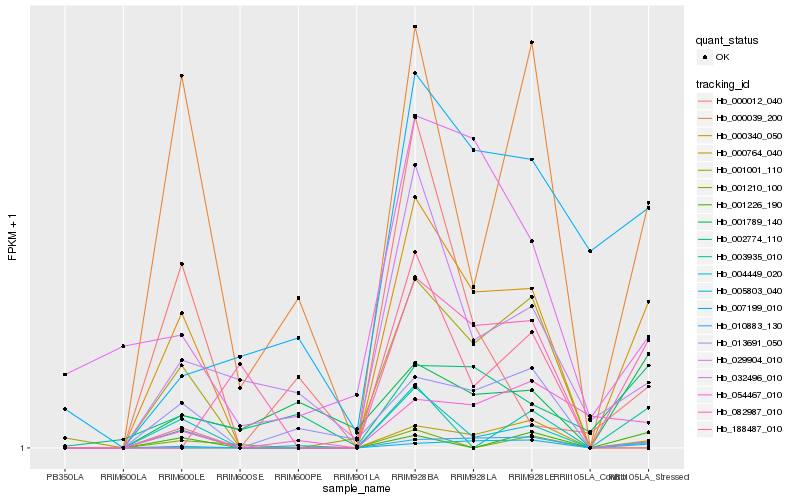
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_050 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_K033901, partial [Eucalyptus grandis] |
| 2 |
Hb_005803_040 |
0.2418262192 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_004449_020 |
0.2486088805 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type isoform X3 [Elaeis guineensis] |
| 4 |
Hb_000039_200 |
0.2757007986 |
- |
- |
hypothetical protein JCGZ_21346 [Jatropha curcas] |
| 5 |
Hb_003935_010 |
0.3144657255 |
- |
- |
- |
| 6 |
Hb_029904_010 |
0.3172033568 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 7 |
Hb_001001_110 |
0.3211743966 |
- |
- |
- |
| 8 |
Hb_010883_130 |
0.3211758461 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 9 |
Hb_013691_050 |
0.3271959796 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 10 |
Hb_000764_040 |
0.3293802954 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 11 |
Hb_032496_010 |
0.3352791268 |
- |
- |
hypothetical protein POPTR_0012s10850g [Populus trichocarpa] |
| 12 |
Hb_054467_010 |
0.3446258484 |
- |
- |
- |
| 13 |
Hb_002774_110 |
0.3487250649 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_001226_190 |
0.3510141138 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 15 |
Hb_001789_140 |
0.3539554925 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Citrus sinensis] |
| 16 |
Hb_007199_010 |
0.3543185685 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |
| 17 |
Hb_001210_100 |
0.3566070176 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 18 |
Hb_188487_010 |
0.3574120022 |
- |
- |
hypothetical protein JCGZ_24382 [Jatropha curcas] |
| 19 |
Hb_082987_010 |
0.3641755892 |
- |
- |
hypothetical protein CICLE_v10030423mg [Citrus clementina] |
| 20 |
Hb_000012_040 |
0.3674292817 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |