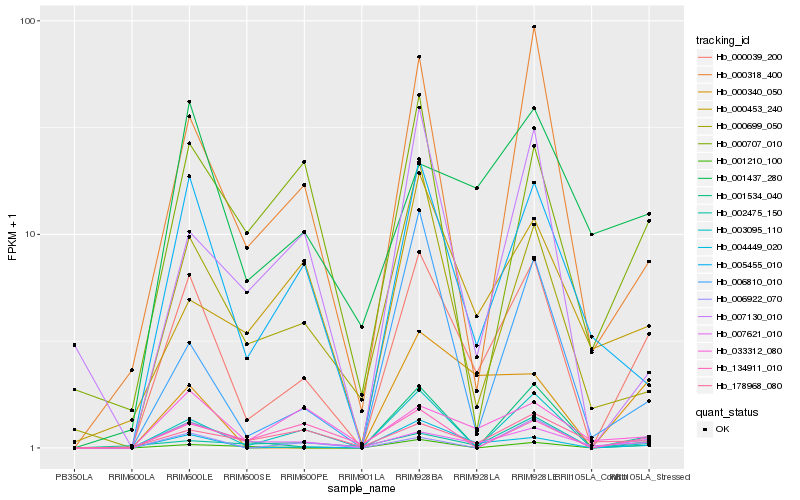
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000039_200 |
0.0 |
- |
- |
hypothetical protein JCGZ_21346 [Jatropha curcas] |
| 2 |
Hb_007621_010 |
0.2184360231 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004449_020 |
0.2241157269 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type isoform X3 [Elaeis guineensis] |
| 4 |
Hb_005455_010 |
0.2301284731 |
- |
- |
PREDICTED: probable acyl-activating enzyme 18, peroxisomal isoform X1 [Jatropha curcas] |
| 5 |
Hb_000318_400 |
0.2441988086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003095_110 |
0.2484138446 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 7 |
Hb_000699_050 |
0.2588879604 |
- |
- |
- |
| 8 |
Hb_006922_070 |
0.2636818868 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 9 |
Hb_001210_100 |
0.2702662762 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 10 |
Hb_033312_080 |
0.2705001926 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 11 |
Hb_001437_280 |
0.2728091803 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 12 |
Hb_000340_050 |
0.2757007986 |
- |
- |
hypothetical protein EUGRSUZ_K033901, partial [Eucalyptus grandis] |
| 13 |
Hb_002475_150 |
0.2768505539 |
- |
- |
PREDICTED: importin subunit beta-3-like [Jatropha curcas] |
| 14 |
Hb_178968_080 |
0.2789930716 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006810_010 |
0.2805135085 |
- |
- |
hypothetical protein B456_012G018200 [Gossypium raimondii] |
| 16 |
Hb_000707_010 |
0.2808848783 |
- |
- |
hypothetical protein JCGZ_25098 [Jatropha curcas] |
| 17 |
Hb_001534_040 |
0.2823552789 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 18 |
Hb_007130_010 |
0.2847690808 |
- |
- |
- |
| 19 |
Hb_000453_240 |
0.2857832548 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 20 |
Hb_134911_010 |
0.285844607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |