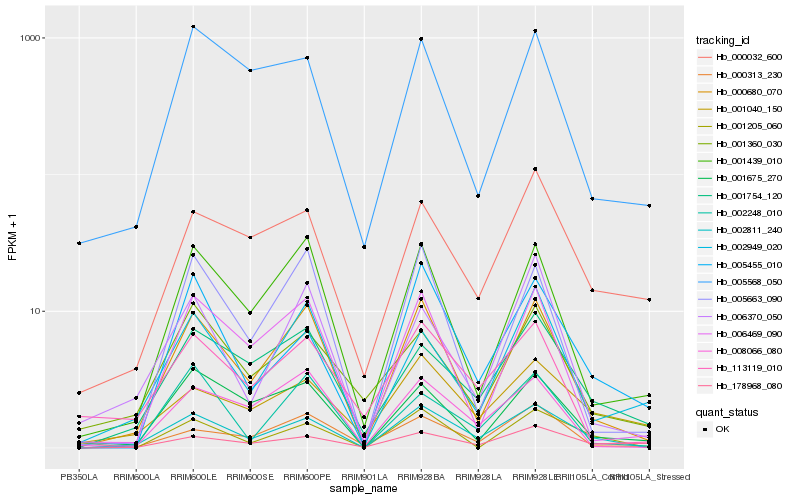
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000680_070 |
0.1414670572 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005455_010 |
0.1718278421 |
- |
- |
PREDICTED: probable acyl-activating enzyme 18, peroxisomal isoform X1 [Jatropha curcas] |
| 4 |
Hb_001754_120 |
0.1830043027 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 5 |
Hb_001040_150 |
0.1871701611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_178968_080 |
0.1972256462 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000032_600 |
0.198536854 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 8 |
Hb_001439_010 |
0.1993315825 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 9 |
Hb_000313_230 |
0.2006619257 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 10 |
Hb_008066_080 |
0.2065671512 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 11 |
Hb_113119_010 |
0.2093816217 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001675_270 |
0.2096893613 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 13 |
Hb_001205_060 |
0.2106361356 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 14 |
Hb_006469_090 |
0.2109211994 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 15 |
Hb_002949_020 |
0.212999122 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 16 |
Hb_002248_010 |
0.2130129939 |
- |
- |
PREDICTED: uncharacterized protein LOC105124180 [Populus euphratica] |
| 17 |
Hb_005568_050 |
0.2130483457 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 18 |
Hb_006370_050 |
0.2132565107 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_001360_030 |
0.2152815568 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 20 |
Hb_005663_090 |
0.2159207312 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |