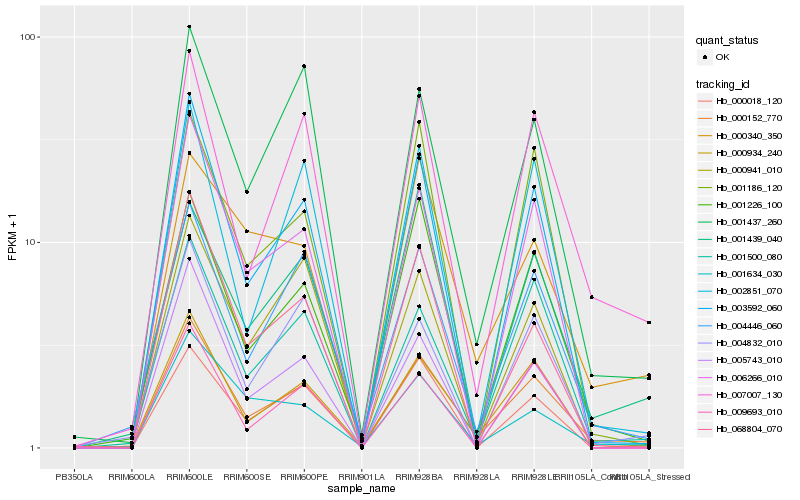
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_770 |
0.0 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 2 |
Hb_000934_240 |
0.106017629 |
- |
- |
PREDICTED: peroxidase 13 [Jatropha curcas] |
| 3 |
Hb_007007_130 |
0.1151395586 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 4 |
Hb_004446_060 |
0.1259623513 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 5 |
Hb_003592_060 |
0.1260090397 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 6 |
Hb_001226_100 |
0.1270288482 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001500_080 |
0.1315489198 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001437_260 |
0.1383616344 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 9 |
Hb_006266_010 |
0.1437268247 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002851_070 |
0.1470236775 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 11 |
Hb_068804_070 |
0.1500112799 |
- |
- |
hypothetical protein JCGZ_12627 [Jatropha curcas] |
| 12 |
Hb_001634_030 |
0.1523616643 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |
| 13 |
Hb_000340_350 |
0.1524389986 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000018_120 |
0.1578676355 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001186_120 |
0.1605266121 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 16 |
Hb_005743_010 |
0.1610554954 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 17 |
Hb_001439_040 |
0.1623858281 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004832_010 |
0.1631276679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000941_010 |
0.1645392827 |
- |
- |
hypothetical protein RCOM_0504460 [Ricinus communis] |
| 20 |
Hb_009693_010 |
0.1645624677 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |