| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
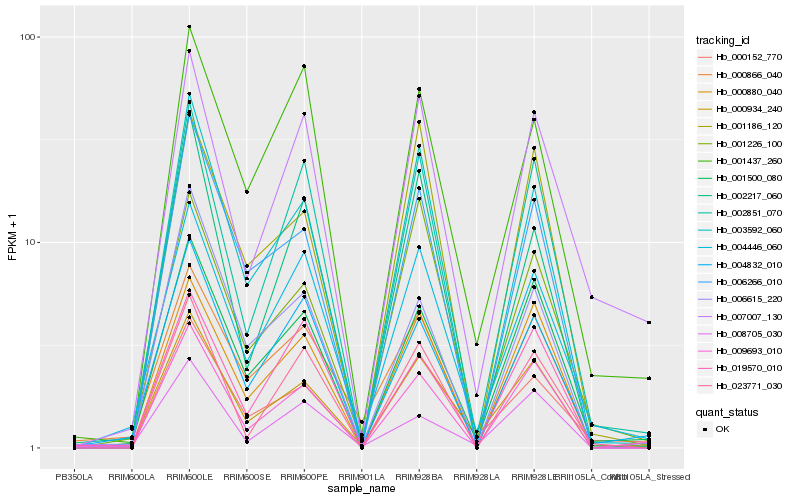
Hb_000934_240 |
0.0 |
- |
- |
PREDICTED: peroxidase 13 [Jatropha curcas] |
| 2 |
Hb_001500_080 |
0.1031913099 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000152_770 |
0.106017629 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 4 |
Hb_003592_060 |
0.120453436 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 5 |
Hb_009693_010 |
0.1214768051 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 6 |
Hb_004446_060 |
0.1228485255 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 7 |
Hb_002851_070 |
0.1290707841 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 8 |
Hb_006266_010 |
0.1301636634 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000880_040 |
0.1348931099 |
- |
- |
- |
| 10 |
Hb_001226_100 |
0.1402675464 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_007007_130 |
0.1440028247 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 12 |
Hb_000866_040 |
0.1473521732 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 13 |
Hb_004832_010 |
0.1504472942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008705_030 |
0.1516880488 |
- |
- |
hypothetical protein JCGZ_13017 [Jatropha curcas] |
| 15 |
Hb_002217_060 |
0.1536078655 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 16 |
Hb_001437_260 |
0.1549555952 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 17 |
Hb_001186_120 |
0.1558915365 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 18 |
Hb_006615_220 |
0.1568835768 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_019570_010 |
0.1570183894 |
- |
- |
lyase, putative [Ricinus communis] |
| 20 |
Hb_023771_030 |
0.1583098028 |
- |
- |
unnamed protein product [Vitis vinifera] |