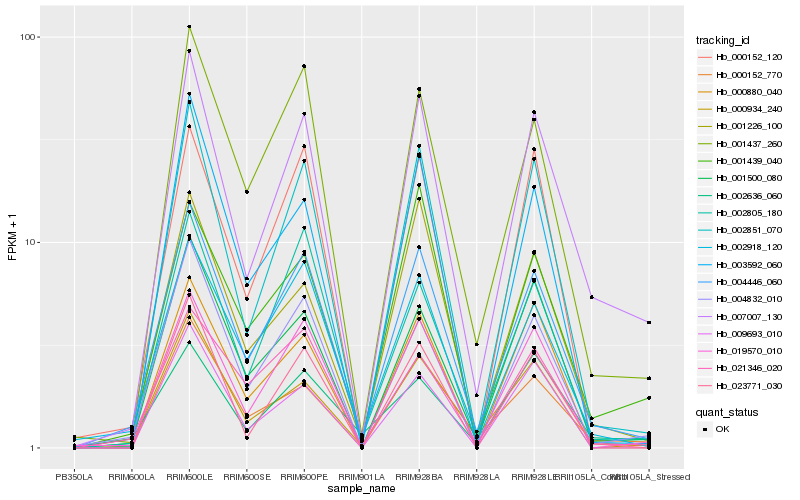
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007007_130 |
0.0 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 2 |
Hb_004446_060 |
0.0995400972 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 3 |
Hb_002851_070 |
0.103642561 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 4 |
Hb_000152_770 |
0.1151395586 |
- |
- |
PREDICTED: dynein light chain, cytoplasmic-like [Jatropha curcas] |
| 5 |
Hb_023771_030 |
0.1264193289 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_001500_080 |
0.1305127741 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001437_260 |
0.1356772051 |
- |
- |
PREDICTED: protein FAM98B-like [Jatropha curcas] |
| 8 |
Hb_004832_010 |
0.1406893131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000934_240 |
0.1440028247 |
- |
- |
PREDICTED: peroxidase 13 [Jatropha curcas] |
| 10 |
Hb_001226_100 |
0.1446454364 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_001439_040 |
0.152219444 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_019570_010 |
0.1532949976 |
- |
- |
lyase, putative [Ricinus communis] |
| 13 |
Hb_000152_120 |
0.1555922522 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 14 |
Hb_002918_120 |
0.1560175981 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_009693_010 |
0.1572149879 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 16 |
Hb_003592_060 |
0.1575741035 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 17 |
Hb_000880_040 |
0.1579530787 |
- |
- |
- |
| 18 |
Hb_002636_060 |
0.1581925175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_021346_020 |
0.1587415164 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 20 |
Hb_002805_180 |
0.1593101011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |