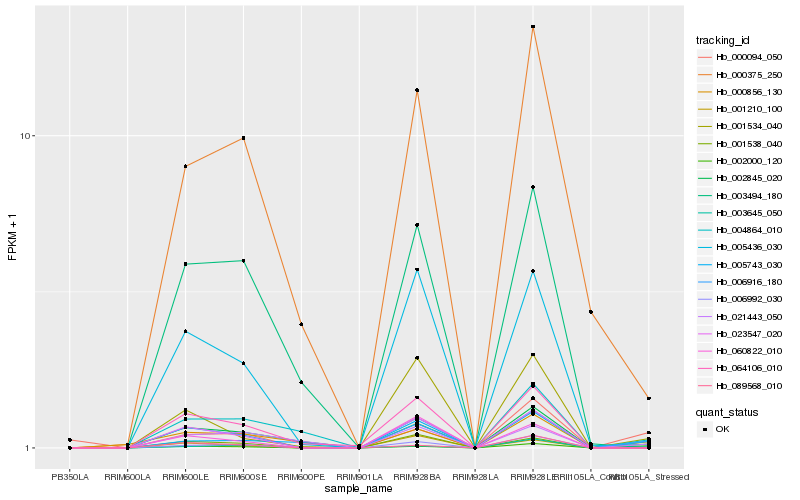
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_180 |
0.0 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 2 |
Hb_002000_120 |
0.1781454805 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_023547_020 |
0.1880267273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005436_030 |
0.192084568 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74E1-like [Jatropha curcas] |
| 5 |
Hb_001210_100 |
0.1957272545 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 6 |
Hb_003645_050 |
0.2076015126 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_001534_040 |
0.2150100058 |
- |
- |
hypothetical protein JCGZ_08436 [Jatropha curcas] |
| 8 |
Hb_000856_130 |
0.2207264207 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_064106_010 |
0.2240828268 |
- |
- |
PREDICTED: uncharacterized protein LOC104878249 [Vitis vinifera] |
| 10 |
Hb_006992_030 |
0.2242907514 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_004864_010 |
0.2286665379 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 12 |
Hb_001538_040 |
0.2294658365 |
- |
- |
PREDICTED: uncharacterized protein LOC104228304 [Nicotiana sylvestris] |
| 13 |
Hb_021443_050 |
0.2344450408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000094_050 |
0.2345639162 |
- |
- |
- |
| 15 |
Hb_000375_250 |
0.2353669839 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 16 |
Hb_005743_030 |
0.2390030576 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 17 |
Hb_003494_180 |
0.2400730841 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 18 |
Hb_089568_010 |
0.2437601377 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 19 |
Hb_060822_010 |
0.2457155215 |
- |
- |
- |
| 20 |
Hb_002845_020 |
0.2458611721 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |