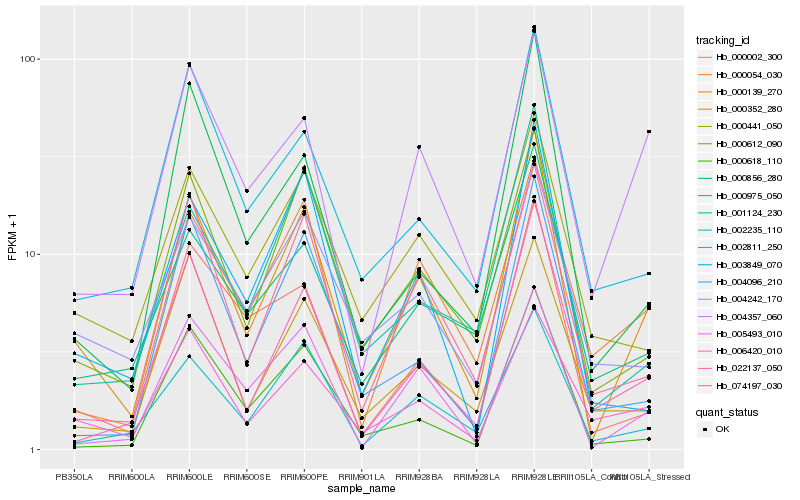
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_270 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s14890g [Populus trichocarpa] |
| 2 |
Hb_000612_090 |
0.1369503531 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 3 |
Hb_022137_050 |
0.1439060931 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 4 |
Hb_002235_110 |
0.1448159097 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 5 |
Hb_074197_030 |
0.1485710843 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_005493_010 |
0.1509945292 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_006420_010 |
0.1522069807 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 8 |
Hb_003849_070 |
0.1559486316 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 9 |
Hb_001124_230 |
0.1565993878 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000975_050 |
0.1574107875 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000054_030 |
0.1592697046 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 12 |
Hb_000002_300 |
0.1601786069 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 13 |
Hb_000856_280 |
0.1615782485 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 14 |
Hb_002811_250 |
0.1622974808 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000441_050 |
0.1638069749 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 16 |
Hb_000352_280 |
0.1656298854 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 17 |
Hb_004357_060 |
0.1658301506 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 18 |
Hb_000618_110 |
0.1685091162 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 19 |
Hb_004096_210 |
0.168580889 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004242_170 |
0.1687687698 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |