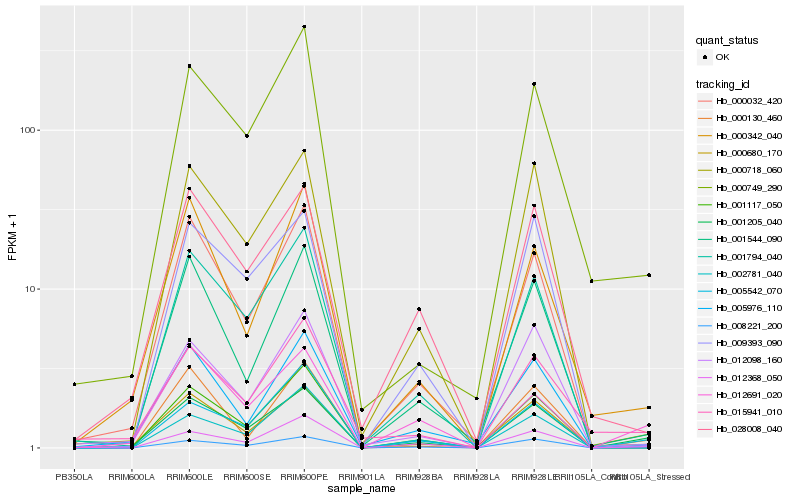
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_040 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_005542_070 |
0.1391121534 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 3 |
Hb_012691_020 |
0.1425498969 |
- |
- |
PREDICTED: MFP1 attachment factor 1-like [Jatropha curcas] |
| 4 |
Hb_002781_040 |
0.1477961916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000718_060 |
0.1479197208 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_001544_090 |
0.1555167566 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 7 |
Hb_012368_050 |
0.1559167786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000749_290 |
0.1571911759 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 9 |
Hb_000032_420 |
0.1573641302 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 10 |
Hb_028008_040 |
0.1589406815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001794_040 |
0.1613895092 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009393_090 |
0.161443962 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000680_170 |
0.1617049609 |
- |
- |
hypothetical protein JCGZ_24789 [Jatropha curcas] |
| 14 |
Hb_001117_050 |
0.162290637 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 15 |
Hb_015941_010 |
0.1627639534 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 16 |
Hb_008221_200 |
0.1637052353 |
- |
- |
PREDICTED: protein trichome birefringence-like 41 [Jatropha curcas] |
| 17 |
Hb_012098_160 |
0.1638872795 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 18 |
Hb_000130_460 |
0.1641272344 |
- |
- |
hypothetical protein JCGZ_15296 [Jatropha curcas] |
| 19 |
Hb_000342_040 |
0.1645115981 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 20 |
Hb_005976_110 |
0.1645689518 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |