| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
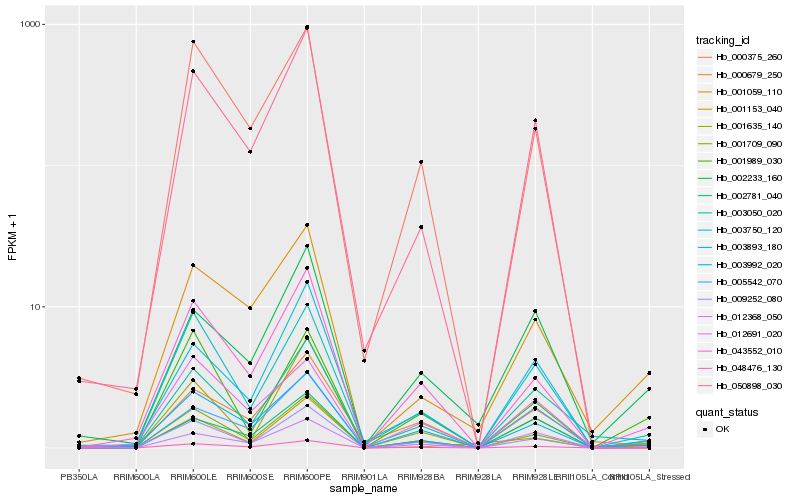
Hb_048476_130 |
0.0 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 2 |
Hb_001059_110 |
0.1235866524 |
- |
- |
PREDICTED: uncharacterized protein LOC105649457 [Jatropha curcas] |
| 3 |
Hb_003992_020 |
0.1266187392 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 4 |
Hb_005542_070 |
0.137906119 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 5 |
Hb_012691_020 |
0.1436683692 |
- |
- |
PREDICTED: MFP1 attachment factor 1-like [Jatropha curcas] |
| 6 |
Hb_002781_040 |
0.1541053531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003893_180 |
0.1554675822 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 8 |
Hb_001989_030 |
0.1563059964 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 9 |
Hb_002233_160 |
0.1590740305 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 10 |
Hb_003050_020 |
0.1617312285 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 11 |
Hb_001709_090 |
0.1618374768 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 12 |
Hb_003750_120 |
0.1652519798 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 13 |
Hb_050898_030 |
0.1664434678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000679_250 |
0.1709800709 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001153_040 |
0.1721569593 |
- |
- |
PREDICTED: uncharacterized protein LOC105128973 [Populus euphratica] |
| 16 |
Hb_009252_080 |
0.1725823333 |
- |
- |
Polygalacturonase-1 non-catalytic subunit beta precursor, putative [Ricinus communis] |
| 17 |
Hb_000375_260 |
0.1727287353 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 18 |
Hb_001635_140 |
0.1728464145 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_043552_010 |
0.1736709968 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 20 |
Hb_012368_050 |
0.1753303521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |