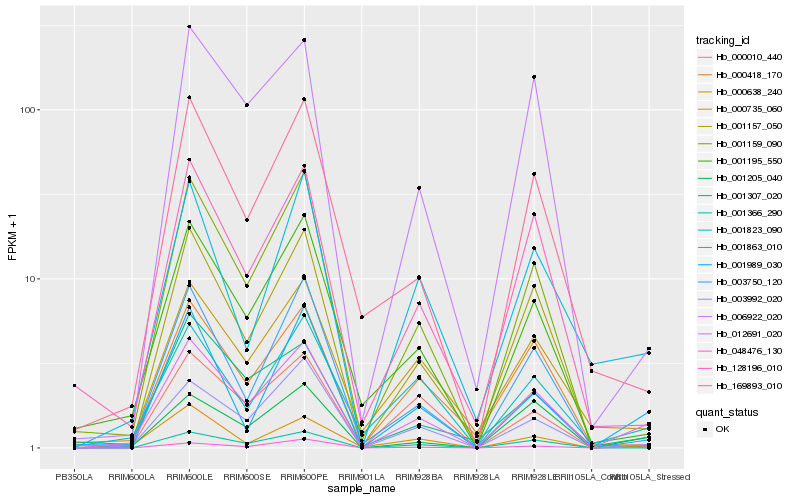
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012691_020 |
0.0 |
- |
- |
PREDICTED: MFP1 attachment factor 1-like [Jatropha curcas] |
| 2 |
Hb_003992_020 |
0.13892222 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 3 |
Hb_001205_040 |
0.1425498969 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_048476_130 |
0.1436683692 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 5 |
Hb_003750_120 |
0.1553258487 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 6 |
Hb_001195_550 |
0.1567399605 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001159_090 |
0.1587082821 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 8 |
Hb_001823_090 |
0.1591616046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006922_020 |
0.161582182 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 10 |
Hb_001366_290 |
0.1623955552 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000638_240 |
0.1635665768 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Prunus mume] |
| 12 |
Hb_001989_030 |
0.1648651138 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 13 |
Hb_128196_010 |
0.1659764362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000735_060 |
0.1664088259 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 15 |
Hb_001307_020 |
0.1680057974 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 16 |
Hb_000418_170 |
0.1689492274 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 17 |
Hb_001157_050 |
0.1692012977 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 18 |
Hb_169893_010 |
0.1698309675 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 19 |
Hb_000010_440 |
0.1702555451 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 20 |
Hb_001863_010 |
0.1705029961 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |