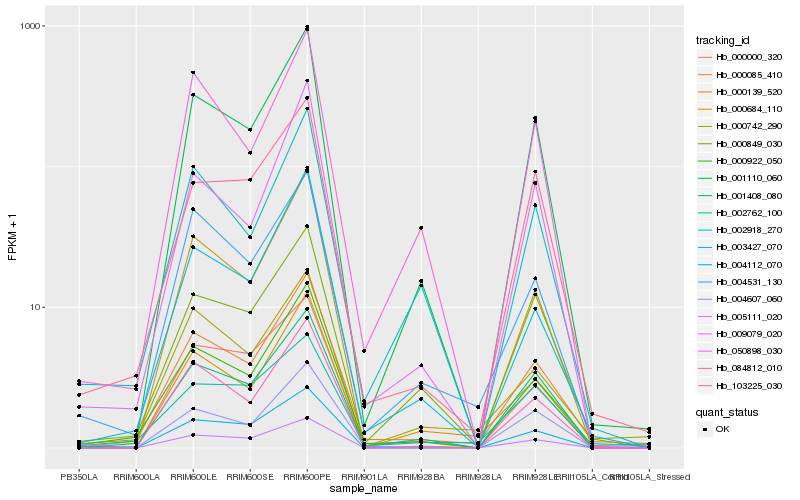
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_410 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 2 |
Hb_000922_050 |
0.0606122782 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 3 |
Hb_001110_060 |
0.0913930501 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 4 |
Hb_001408_080 |
0.0990498855 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 5 |
Hb_000742_290 |
0.1071653139 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 6 |
Hb_003427_070 |
0.1099061476 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 7 |
Hb_004531_130 |
0.1130576929 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_103225_030 |
0.1146808575 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000139_520 |
0.1148947612 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 10 |
Hb_000684_110 |
0.1169108305 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 11 |
Hb_084812_010 |
0.1190702246 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 12 |
Hb_000849_030 |
0.1190832061 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 13 |
Hb_005111_020 |
0.1208320039 |
- |
- |
hypothetical protein CISIN_1g0417821mg, partial [Citrus sinensis] |
| 14 |
Hb_002918_270 |
0.1211131228 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 15 |
Hb_009079_020 |
0.1240642752 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 16 |
Hb_004112_070 |
0.124485849 |
- |
- |
- |
| 17 |
Hb_050898_030 |
0.1246511288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002762_100 |
0.1255671929 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000000_320 |
0.1262274263 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004607_060 |
0.1265272858 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |