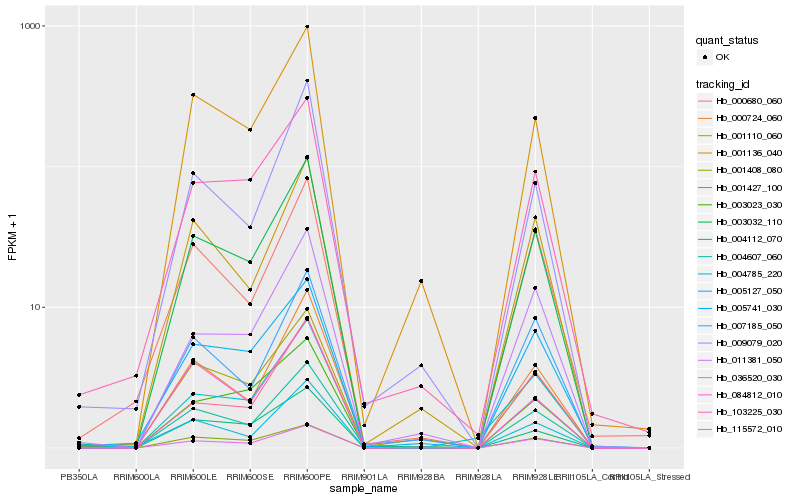
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003032_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_036520_030 |
0.086637864 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 3 |
Hb_001110_060 |
0.0878158765 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 4 |
Hb_005741_030 |
0.0892130368 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 5 |
Hb_000724_060 |
0.0937656353 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 6 |
Hb_001408_080 |
0.094149808 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 7 |
Hb_001136_040 |
0.0946771321 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_004785_220 |
0.0993416067 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.2 [Populus euphratica] |
| 9 |
Hb_115572_010 |
0.1013499172 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 10 |
Hb_004112_070 |
0.1015837888 |
- |
- |
- |
| 11 |
Hb_103225_030 |
0.1018477231 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_004607_060 |
0.1046516411 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 13 |
Hb_011381_050 |
0.1078238835 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 14 |
Hb_001427_100 |
0.1118299228 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 15 |
Hb_007185_050 |
0.1136974326 |
- |
- |
hypothetical protein JCGZ_22475 [Jatropha curcas] |
| 16 |
Hb_000680_060 |
0.1172110529 |
- |
- |
hypothetical protein POPTR_0001s20790g [Populus trichocarpa] |
| 17 |
Hb_009079_020 |
0.1172488214 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 18 |
Hb_084812_010 |
0.1172597345 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 19 |
Hb_005127_050 |
0.1193654708 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 20 |
Hb_003023_030 |
0.1195237528 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |