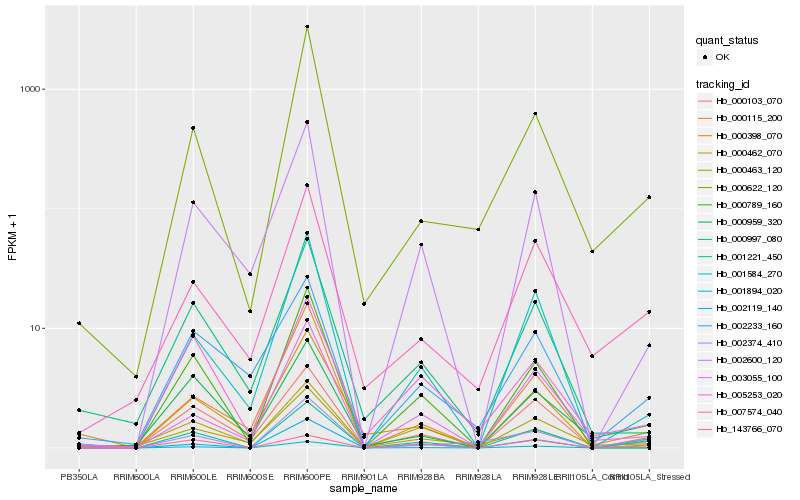
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_450 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002374_410 |
0.17316804 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000398_070 |
0.1800630274 |
- |
- |
hypothetical protein VITISV_011880 [Vitis vinifera] |
| 4 |
Hb_001894_020 |
0.1845667356 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP630 [Jatropha curcas] |
| 5 |
Hb_000103_070 |
0.1975428015 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 6 |
Hb_002119_140 |
0.2055936064 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 7 |
Hb_001584_270 |
0.2097311303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000622_120 |
0.2112648196 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 9 |
Hb_000997_080 |
0.212520205 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003055_100 |
0.2127172196 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Jatropha curcas] |
| 11 |
Hb_000115_200 |
0.2165448216 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 12 |
Hb_000959_320 |
0.2169862447 |
- |
- |
PREDICTED: uncharacterized protein LOC105778325 [Gossypium raimondii] |
| 13 |
Hb_007574_040 |
0.217919392 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 14 |
Hb_000462_070 |
0.2182462976 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 15 |
Hb_002233_160 |
0.2190182867 |
- |
- |
PREDICTED: uncharacterized protein LOC105633094 [Jatropha curcas] |
| 16 |
Hb_002600_120 |
0.2199285829 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 17 |
Hb_143766_070 |
0.2202825077 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 18 |
Hb_000789_160 |
0.222932883 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 19 |
Hb_005253_020 |
0.226868834 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 20 |
Hb_000463_120 |
0.2277461858 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |