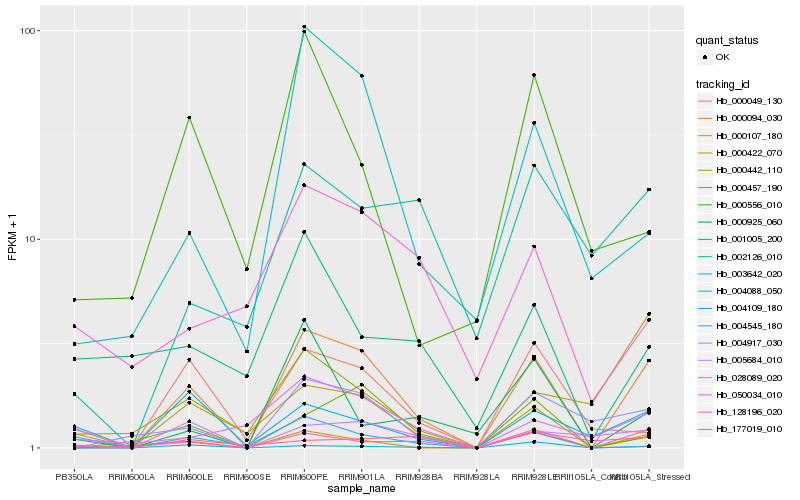
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000422_070 |
0.2496713348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000107_180 |
0.2564597231 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 4 |
Hb_004917_030 |
0.2770745005 |
- |
- |
- |
| 5 |
Hb_004109_180 |
0.2818593798 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003642_020 |
0.2930020207 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 7 |
Hb_000457_190 |
0.2951787511 |
- |
- |
- |
| 8 |
Hb_004088_050 |
0.298235223 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 9 |
Hb_028089_020 |
0.2996207441 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 10 |
Hb_128196_020 |
0.3016650986 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Nelumbo nucifera] |
| 11 |
Hb_177019_010 |
0.3021844912 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 12 |
Hb_004545_180 |
0.308114019 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_000049_130 |
0.3132782915 |
- |
- |
PREDICTED: uncharacterized protein LOC104879855 [Vitis vinifera] |
| 14 |
Hb_005684_010 |
0.3185220859 |
- |
- |
PREDICTED: uncharacterized protein LOC104216799 [Nicotiana sylvestris] |
| 15 |
Hb_002126_010 |
0.3252292456 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 16 |
Hb_000925_060 |
0.3258224641 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |
| 17 |
Hb_001005_200 |
0.3294600013 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_050034_010 |
0.331287723 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 19 |
Hb_000556_010 |
0.3331350288 |
- |
- |
glycerate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_000442_110 |
0.3332958357 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |