| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
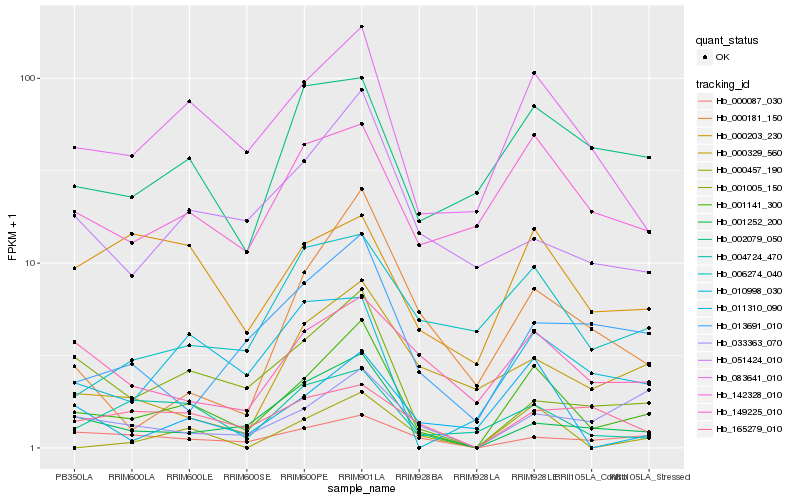
Hb_001141_300 |
0.0 |
- |
- |
- |
| 2 |
Hb_083641_010 |
0.1976018792 |
- |
- |
SECY isoform 1 [Theobroma cacao] |
| 3 |
Hb_001252_200 |
0.2066556934 |
- |
- |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 4 |
Hb_001005_150 |
0.2224480953 |
- |
- |
- |
| 5 |
Hb_000087_030 |
0.2355276495 |
- |
- |
hypothetical protein JCGZ_02243 [Jatropha curcas] |
| 6 |
Hb_004724_470 |
0.2438838796 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 7 |
Hb_142328_010 |
0.2448830954 |
- |
- |
PREDICTED: uncharacterized protein LOC105630513 isoform X1 [Jatropha curcas] |
| 8 |
Hb_011310_090 |
0.2477227542 |
- |
- |
PREDICTED: putative ATPase N2B isoform X3 [Vitis vinifera] |
| 9 |
Hb_013691_010 |
0.24806413 |
- |
- |
PREDICTED: uncharacterized protein LOC103724099, partial [Phoenix dactylifera] |
| 10 |
Hb_051424_010 |
0.2486439446 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 11 |
Hb_010998_030 |
0.2489084548 |
- |
- |
- |
| 12 |
Hb_033363_070 |
0.2498270576 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methionine aminopeptidase 1B, chloroplastic, partial [Populus euphratica] |
| 13 |
Hb_002079_050 |
0.2511125718 |
- |
- |
hypothetical protein B456_004G182300 [Gossypium raimondii] |
| 14 |
Hb_149225_010 |
0.2552331597 |
- |
- |
DNA-directed RNA polymerase II subunit rpb4, putative isoform 4, partial [Theobroma cacao] |
| 15 |
Hb_000181_150 |
0.2611217394 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 16 |
Hb_000457_190 |
0.2620941257 |
- |
- |
- |
| 17 |
Hb_000203_230 |
0.2646629743 |
- |
- |
PREDICTED: probable inactive dual specificity protein phosphatase-like At4g18593 [Jatropha curcas] |
| 18 |
Hb_165279_010 |
0.2655937048 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 19 |
Hb_000329_560 |
0.2656262442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006274_040 |
0.2681121524 |
- |
- |
PREDICTED: DENN domain and WD repeat-containing protein SCD1-like [Fragaria vesca subsp. vesca] |