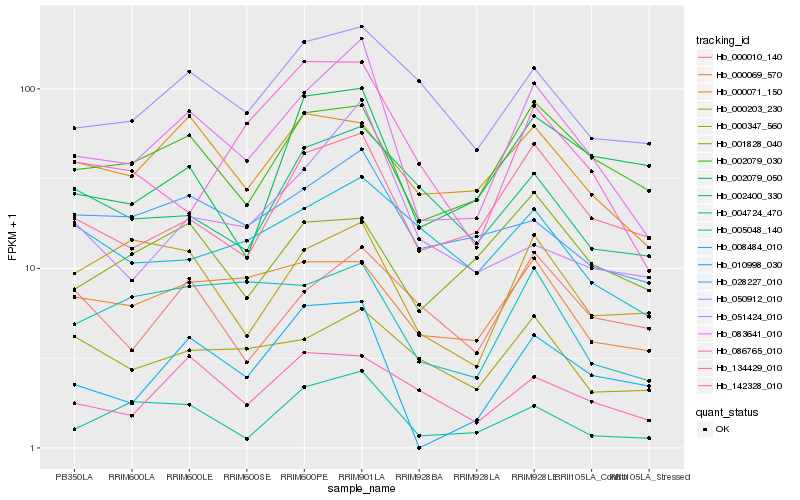
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083641_010 |
0.0 |
- |
- |
SECY isoform 1 [Theobroma cacao] |
| 2 |
Hb_142328_010 |
0.1459096971 |
- |
- |
PREDICTED: uncharacterized protein LOC105630513 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002079_030 |
0.1524791464 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 4 |
Hb_050912_010 |
0.1716407212 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_004724_470 |
0.1730096124 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 6 |
Hb_010998_030 |
0.1734660743 |
- |
- |
- |
| 7 |
Hb_005048_140 |
0.1734687283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_028227_010 |
0.1750665362 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 9 |
Hb_000071_150 |
0.1778690444 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 10 |
Hb_002079_050 |
0.180044083 |
- |
- |
hypothetical protein B456_004G182300 [Gossypium raimondii] |
| 11 |
Hb_000069_570 |
0.1807442949 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_134429_010 |
0.1809393934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000203_230 |
0.1818080126 |
- |
- |
PREDICTED: probable inactive dual specificity protein phosphatase-like At4g18593 [Jatropha curcas] |
| 14 |
Hb_000347_560 |
0.1854084124 |
- |
- |
- |
| 15 |
Hb_002400_330 |
0.1868097717 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_000010_140 |
0.1884771354 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 11, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_051424_010 |
0.191435197 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 18 |
Hb_008484_010 |
0.1951650063 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 19 |
Hb_086765_010 |
0.1967453844 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 20 |
Hb_001828_040 |
0.1972272725 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |