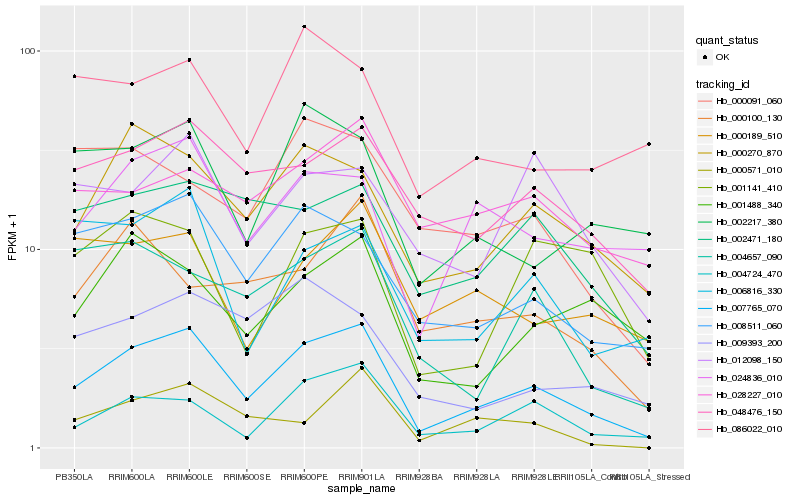
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_070 |
0.0 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |
| 2 |
Hb_008511_060 |
0.1546039068 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 3 |
Hb_000270_870 |
0.1589931303 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 4 |
Hb_004724_470 |
0.1613670446 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 5 |
Hb_048476_150 |
0.1639334196 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_024836_010 |
0.1761693599 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 7 |
Hb_002471_180 |
0.1780788722 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_006816_330 |
0.1814842899 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 9 |
Hb_002217_380 |
0.1831918609 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 10 |
Hb_004657_090 |
0.1858077928 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 11 |
Hb_001488_340 |
0.1861655794 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 12 |
Hb_009393_200 |
0.1895309513 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 13 |
Hb_000189_510 |
0.1901420306 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 14 |
Hb_000571_010 |
0.1908556421 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Gossypium raimondii] |
| 15 |
Hb_012098_150 |
0.1933437867 |
- |
- |
ARC5 protein [Manihot esculenta] |
| 16 |
Hb_000100_130 |
0.1939474569 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 17 |
Hb_028227_010 |
0.1941346154 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 18 |
Hb_001141_410 |
0.1973460036 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 19 |
Hb_000091_060 |
0.1980113617 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 20 |
Hb_086022_010 |
0.1990742709 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |