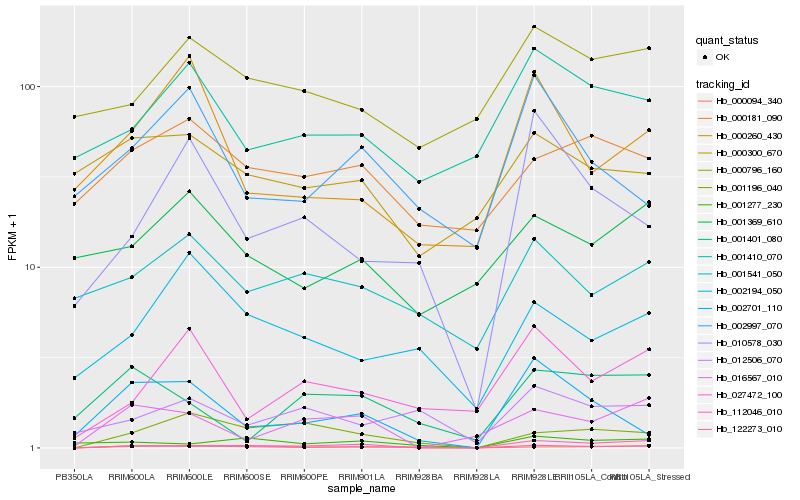
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122273_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000094_340 |
0.2362315348 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 3 |
Hb_002701_110 |
0.2428602491 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1-like [Jatropha curcas] |
| 4 |
Hb_112046_010 |
0.248515981 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 5 |
Hb_001196_040 |
0.2531621704 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 6 |
Hb_016567_010 |
0.2584119854 |
- |
- |
- |
| 7 |
Hb_001277_230 |
0.2659745533 |
- |
- |
- |
| 8 |
Hb_010578_030 |
0.2660088739 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000181_090 |
0.2705217116 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 10 |
Hb_000260_430 |
0.2723006966 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 11 |
Hb_001410_070 |
0.2749476006 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 12 |
Hb_002194_050 |
0.275242543 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002997_070 |
0.2807289651 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 14 |
Hb_027472_100 |
0.2820901014 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 15 |
Hb_001541_050 |
0.2822211834 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 16 |
Hb_012506_070 |
0.2825854826 |
- |
- |
hypothetical protein POPTR_0004s20060g [Populus trichocarpa] |
| 17 |
Hb_000796_160 |
0.2830940757 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000300_670 |
0.2831706676 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001401_080 |
0.2845770848 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001369_610 |
0.2853248824 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |