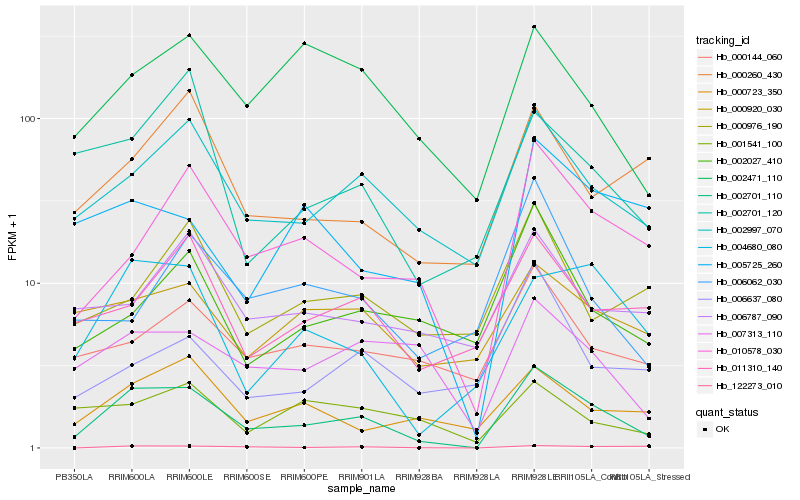
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_110 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1-like [Jatropha curcas] |
| 2 |
Hb_002997_070 |
0.1994030606 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 3 |
Hb_010578_030 |
0.2101792064 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004680_080 |
0.2269244488 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 5 |
Hb_007313_110 |
0.2347430453 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 6 |
Hb_000723_350 |
0.2412724111 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_122273_010 |
0.2428602491 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000144_060 |
0.2443604999 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_011310_140 |
0.2446081141 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |
| 10 |
Hb_002471_110 |
0.2459877263 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 11 |
Hb_002027_410 |
0.2467305426 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000260_430 |
0.2497489669 |
- |
- |
ribosomal protein L17, putative [Ricinus communis] |
| 13 |
Hb_002701_120 |
0.2530514195 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 14 |
Hb_000920_030 |
0.255230446 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_005725_260 |
0.2563695387 |
- |
- |
squalene monooxygenase [Jatropha curcas] |
| 16 |
Hb_006787_090 |
0.2597401005 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 17 |
Hb_006062_030 |
0.2609412387 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001541_100 |
0.2612519103 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 19 |
Hb_000976_190 |
0.2615172362 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 20 |
Hb_006637_080 |
0.2622858698 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |