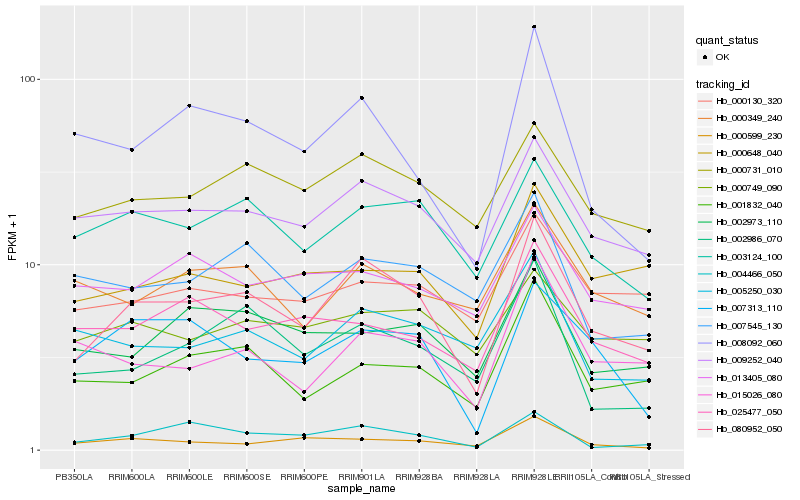
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080952_050 |
0.0 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Nelumbo nucifera] |
| 2 |
Hb_009252_040 |
0.1273234478 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000731_010 |
0.1375520986 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003124_100 |
0.1436033272 |
- |
- |
- |
| 5 |
Hb_001832_040 |
0.1446035999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004466_050 |
0.153826946 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 7 |
Hb_002986_070 |
0.1542889355 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 8 |
Hb_002973_110 |
0.1552224877 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_015026_080 |
0.1585963712 |
- |
- |
- |
| 10 |
Hb_008092_060 |
0.1591418758 |
- |
- |
- |
| 11 |
Hb_000648_040 |
0.1596621256 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_007313_110 |
0.1612113718 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 13 |
Hb_005250_030 |
0.1616061251 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 14 |
Hb_007545_130 |
0.1619851567 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000130_320 |
0.1622713039 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 16 |
Hb_025477_050 |
0.1640339888 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_000349_240 |
0.1640723074 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000749_090 |
0.1650551005 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000599_230 |
0.1696688171 |
- |
- |
- |
| 20 |
Hb_013405_080 |
0.1697715243 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |