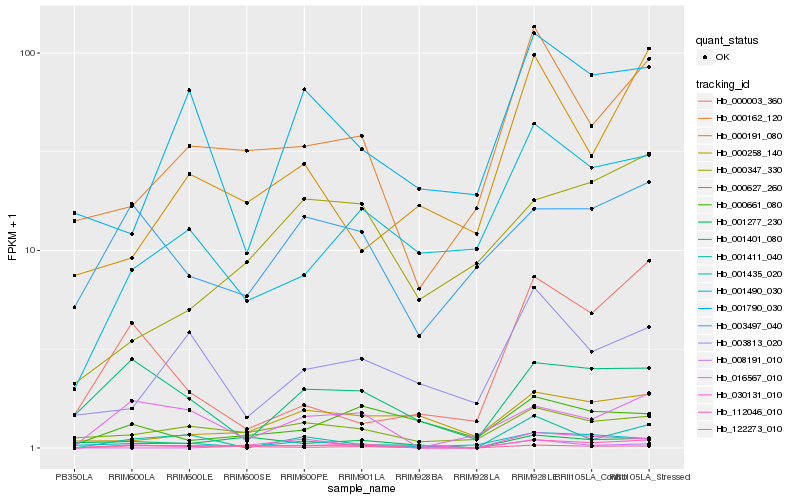
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_112046_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 2 |
Hb_000162_120 |
0.2312268322 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_122273_010 |
0.248515981 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_001490_030 |
0.2698938477 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000627_260 |
0.2733967641 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 6 |
Hb_016567_010 |
0.2754515185 |
- |
- |
- |
| 7 |
Hb_030131_010 |
0.2764263446 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 8 |
Hb_001277_230 |
0.2891889152 |
- |
- |
- |
| 9 |
Hb_008191_010 |
0.292666055 |
- |
- |
- |
| 10 |
Hb_000347_330 |
0.2940150276 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 11 |
Hb_000191_080 |
0.3037336864 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 12 |
Hb_000661_080 |
0.3053242463 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 13 |
Hb_003497_040 |
0.3077811001 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000003_360 |
0.3106027832 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 15 |
Hb_001411_040 |
0.3116026869 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 16 |
Hb_000258_140 |
0.313223005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003813_020 |
0.3140993906 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001435_020 |
0.3162365655 |
- |
- |
hypothetical protein JCGZ_22466 [Jatropha curcas] |
| 19 |
Hb_001401_080 |
0.3181264866 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001790_030 |
0.3186364228 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |