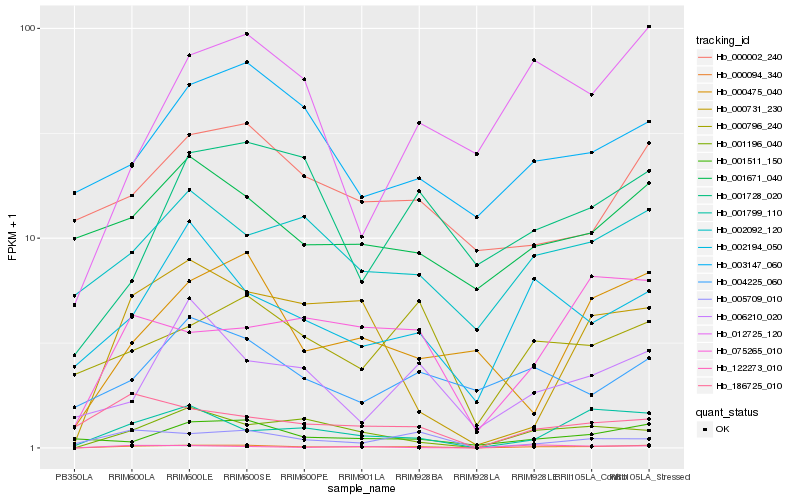
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_340 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 2 |
Hb_001196_040 |
0.207654701 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 3 |
Hb_001799_110 |
0.2175651101 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 4 |
Hb_075265_010 |
0.2186547328 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_005709_010 |
0.224555539 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_002194_050 |
0.2300491339 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000796_240 |
0.2304692513 |
- |
- |
- |
| 8 |
Hb_001511_150 |
0.2351278611 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 9 |
Hb_002092_120 |
0.2360059344 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_122273_010 |
0.2362315348 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_186725_010 |
0.2441150101 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_000731_230 |
0.246019517 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 13 |
Hb_000002_240 |
0.2478062766 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 14 |
Hb_000475_040 |
0.2484164797 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001671_040 |
0.2489962433 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 16 |
Hb_006210_020 |
0.2493351778 |
- |
- |
PREDICTED: calcium sensing receptor, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003147_060 |
0.2505730123 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 18 |
Hb_001728_020 |
0.2507472854 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 19 |
Hb_012725_120 |
0.2509371635 |
- |
- |
PREDICTED: stress enhanced protein 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004225_060 |
0.2515422211 |
- |
- |
hypothetical protein CISIN_1g027511mg [Citrus sinensis] |