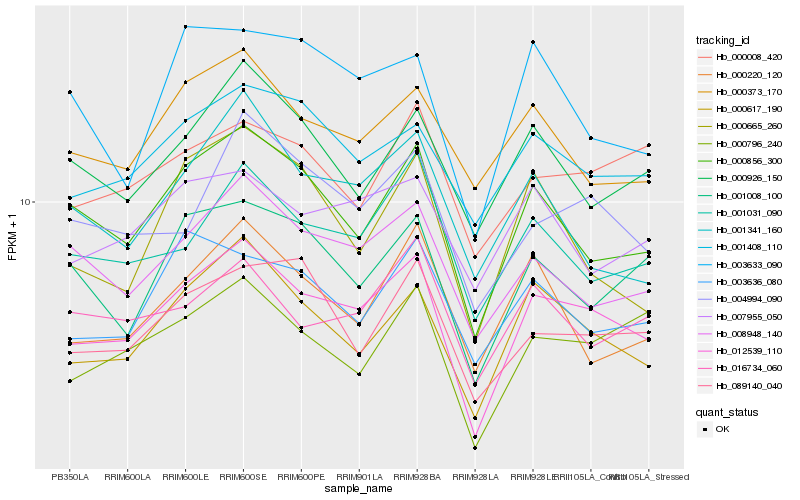
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012539_110 |
0.0 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_000617_190 |
0.0855028602 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |
| 3 |
Hb_000856_300 |
0.0944205276 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 4 |
Hb_000796_240 |
0.1048192373 |
- |
- |
- |
| 5 |
Hb_004994_090 |
0.1093115997 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 6 |
Hb_008948_140 |
0.1099737466 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 7 |
Hb_001031_090 |
0.110356334 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 8 |
Hb_001408_110 |
0.1148273258 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 9 |
Hb_001008_100 |
0.115190491 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 10 |
Hb_089140_040 |
0.1160416769 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 11 |
Hb_000926_150 |
0.1223241746 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_000373_170 |
0.1227356318 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 13 |
Hb_016734_060 |
0.1230741704 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 14 |
Hb_000220_120 |
0.1238048526 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003636_080 |
0.1245044254 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 16 |
Hb_007955_050 |
0.1245342071 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_003633_090 |
0.12500232 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 18 |
Hb_001341_160 |
0.1257872062 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000008_420 |
0.1259582461 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000665_260 |
0.126283202 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |