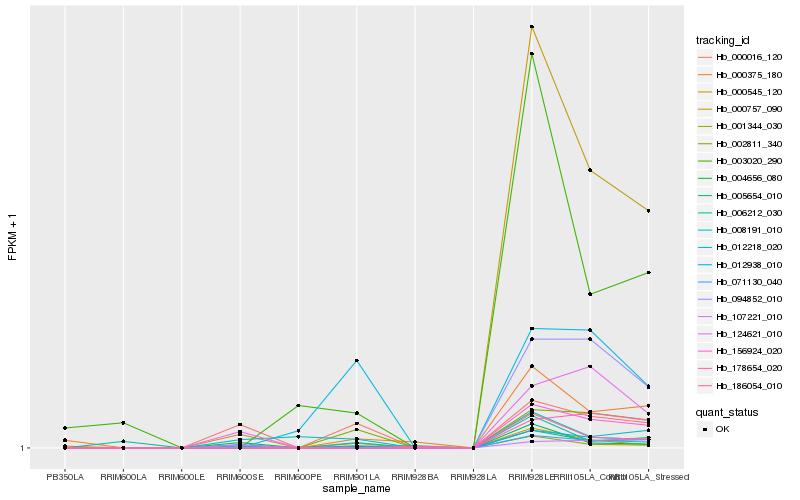
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012938_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_005654_010 |
0.1888525816 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_071130_040 |
0.1909013577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000375_180 |
0.1983855353 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 5 |
Hb_002811_340 |
0.2209577006 |
- |
- |
PREDICTED: ammonium transporter 3 member 2-like [Populus euphratica] |
| 6 |
Hb_000016_120 |
0.2259913292 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 7 |
Hb_156924_020 |
0.2285870989 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 8 |
Hb_124621_010 |
0.2292770523 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 9 |
Hb_000757_090 |
0.2292780714 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 3 [Theobroma cacao] |
| 10 |
Hb_000545_120 |
0.2400789173 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_094852_010 |
0.2472264513 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 12 |
Hb_001344_030 |
0.2686365531 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_178654_020 |
0.2713770786 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 14 |
Hb_003020_290 |
0.2818254324 |
- |
- |
hypothetical protein CICLE_v10029104mg [Citrus clementina] |
| 15 |
Hb_006212_030 |
0.2878367489 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 16 |
Hb_186054_010 |
0.2921274592 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_004656_080 |
0.2952877321 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_008191_010 |
0.2961225189 |
- |
- |
- |
| 19 |
Hb_107221_010 |
0.2981937183 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_012218_020 |
0.3046983401 |
- |
- |
hypothetical protein JCGZ_14688 [Jatropha curcas] |