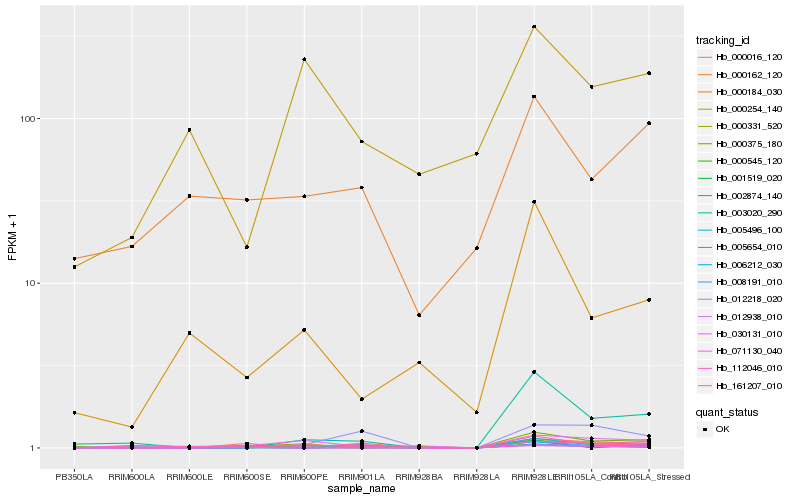
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008191_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_005496_100 |
0.2387443412 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH85-like [Populus euphratica] |
| 3 |
Hb_002874_140 |
0.2580653425 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 11 homolog [Prunus mume] |
| 4 |
Hb_161207_010 |
0.2692333881 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 5 |
Hb_000016_120 |
0.2904573094 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 6 |
Hb_112046_010 |
0.292666055 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 7 |
Hb_012938_010 |
0.2961225189 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_000331_520 |
0.3063985641 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 9 |
Hb_030131_010 |
0.3090085582 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal, partial [Jatropha curcas] |
| 10 |
Hb_001519_020 |
0.3145148085 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 11 |
Hb_000162_120 |
0.3201722351 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_003020_290 |
0.3221843247 |
- |
- |
hypothetical protein CICLE_v10029104mg [Citrus clementina] |
| 13 |
Hb_005654_010 |
0.3224459563 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000375_180 |
0.3243488603 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 15 |
Hb_000545_120 |
0.33305674 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_012218_020 |
0.334595445 |
- |
- |
hypothetical protein JCGZ_14688 [Jatropha curcas] |
| 17 |
Hb_000184_030 |
0.3439225819 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006212_030 |
0.353107282 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 19 |
Hb_071130_040 |
0.3547715563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000254_140 |
0.3550585195 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic [Jatropha curcas] |