| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
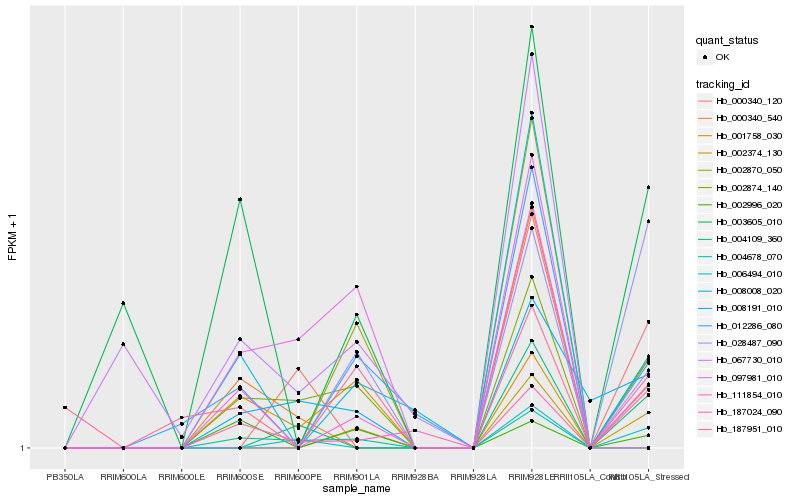
| 1 |
Hb_002874_140 |
0.0 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 11 homolog [Prunus mume] |
| 2 |
Hb_008191_010 |
0.2580653425 |
- |
- |
- |
| 3 |
Hb_111854_010 |
0.2884185469 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 4 |
Hb_000340_540 |
0.3099445922 |
- |
- |
PREDICTED: peroxidase 39-like [Jatropha curcas] |
| 5 |
Hb_187024_090 |
0.3161256846 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_008008_020 |
0.3170467715 |
- |
- |
- |
| 7 |
Hb_002996_020 |
0.3260349575 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 8 |
Hb_067730_010 |
0.3271983726 |
- |
- |
hypothetical protein OsJ_21419 [Oryza sativa Japonica Group] |
| 9 |
Hb_012286_080 |
0.3342094884 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 10 |
Hb_004678_070 |
0.3433179522 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 11 |
Hb_097981_010 |
0.3434661873 |
- |
- |
- |
| 12 |
Hb_003605_010 |
0.3531094503 |
- |
- |
- |
| 13 |
Hb_002374_130 |
0.355134318 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 14 |
Hb_001758_030 |
0.3672630904 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 15 |
Hb_002870_050 |
0.3700946612 |
- |
- |
- |
| 16 |
Hb_028487_090 |
0.3752856883 |
- |
- |
- |
| 17 |
Hb_000340_120 |
0.3828788042 |
- |
- |
PREDICTED: uncharacterized protein LOC103406091 [Malus domestica] |
| 18 |
Hb_004109_360 |
0.3832793505 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1-like [Jatropha curcas] |
| 19 |
Hb_187951_010 |
0.3837809262 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 20 |
Hb_006494_010 |
0.3838500707 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |