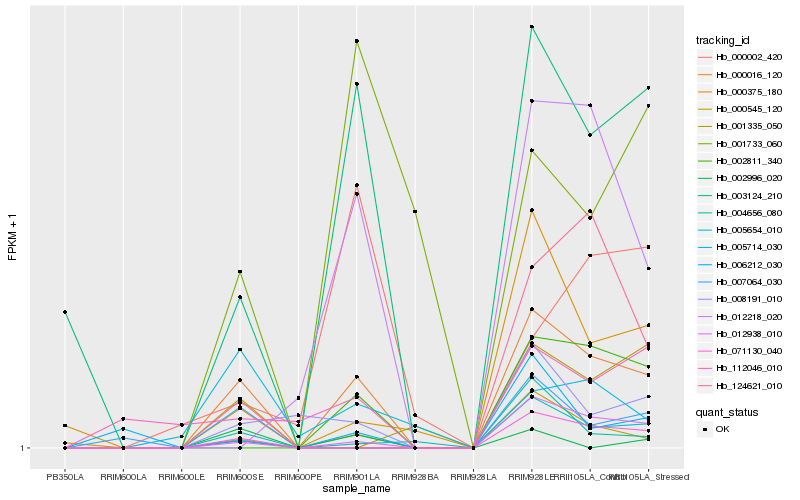
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 2 |
Hb_012938_010 |
0.2259913292 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_003124_210 |
0.2284935846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000545_120 |
0.2459563183 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_000375_180 |
0.2826372048 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 6 |
Hb_005654_010 |
0.2872113418 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_008191_010 |
0.2904573094 |
- |
- |
- |
| 8 |
Hb_002811_340 |
0.2925357999 |
- |
- |
PREDICTED: ammonium transporter 3 member 2-like [Populus euphratica] |
| 9 |
Hb_071130_040 |
0.3015130433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001733_060 |
0.3209004856 |
- |
- |
PREDICTED: uncharacterized protein LOC104431099 [Eucalyptus grandis] |
| 11 |
Hb_012218_020 |
0.3226285705 |
- |
- |
hypothetical protein JCGZ_14688 [Jatropha curcas] |
| 12 |
Hb_005714_030 |
0.3331600726 |
- |
- |
PREDICTED: COBRA-like protein 6 isoform X1 [Gossypium raimondii] |
| 13 |
Hb_006212_030 |
0.3405426091 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 14 |
Hb_112046_010 |
0.3411878524 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 15 |
Hb_000002_420 |
0.3419317465 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1-like [Citrus sinensis] |
| 16 |
Hb_124621_010 |
0.3419451944 |
- |
- |
hypothetical protein L484_019698 [Morus notabilis] |
| 17 |
Hb_007064_030 |
0.3524837668 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 18 |
Hb_001335_050 |
0.3548434835 |
- |
- |
- |
| 19 |
Hb_002996_020 |
0.3609764443 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 20 |
Hb_004656_080 |
0.3624264203 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |