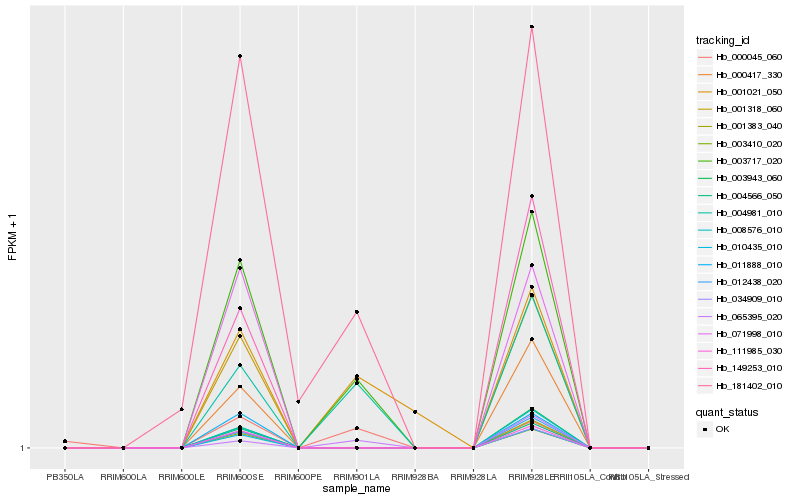
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003717_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_004981_010 |
0.1152623843 |
- |
- |
- |
| 3 |
Hb_181402_010 |
0.1939722363 |
- |
- |
PREDICTED: uncharacterized protein LOC104905886 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_065395_020 |
0.2249474109 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_001021_050 |
0.2288487436 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_000045_060 |
0.2497287758 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_111985_030 |
0.2632664354 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 8 |
Hb_004566_050 |
0.2632665918 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_001318_060 |
0.2637603726 |
- |
- |
contains similarity to reverse transcriptase-like proteins [Arabidopsis thaliana] |
| 10 |
Hb_003410_020 |
0.2640769466 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_034909_010 |
0.2640857819 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_010435_010 |
0.2644005972 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_071998_010 |
0.2675961021 |
- |
- |
PREDICTED: uncharacterized protein LOC105800957 [Gossypium raimondii] |
| 14 |
Hb_011888_010 |
0.2683076424 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 15 |
Hb_000417_330 |
0.2717704771 |
- |
- |
- |
| 16 |
Hb_008576_010 |
0.2733306073 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_149253_010 |
0.2744560345 |
- |
- |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Gossypium arboreum] |
| 18 |
Hb_003943_060 |
0.2746710799 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 19 |
Hb_012438_020 |
0.2750387477 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_001383_040 |
0.2751526455 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |