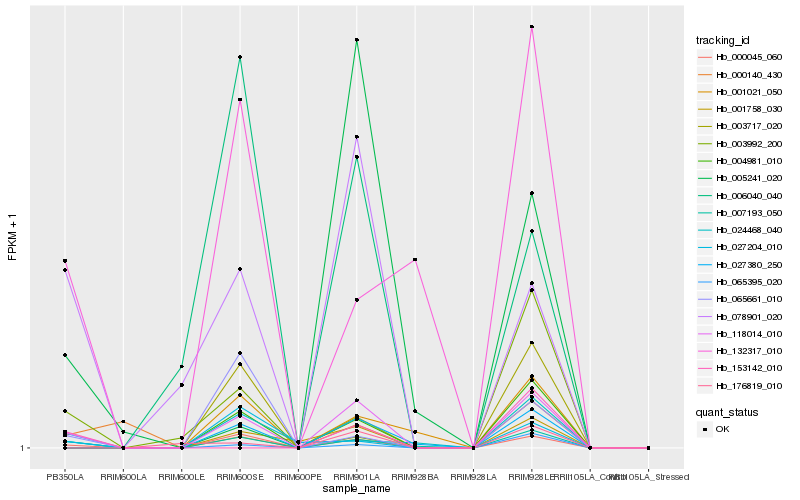
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118014_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104214283 [Nicotiana sylvestris] |
| 2 |
Hb_027380_250 |
0.142125135 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 3 |
Hb_000045_060 |
0.1528918307 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_007193_050 |
0.1863737283 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_004981_010 |
0.25998197 |
- |
- |
- |
| 6 |
Hb_078901_020 |
0.2659244875 |
- |
- |
PREDICTED: uncharacterized protein LOC105648117 [Jatropha curcas] |
| 7 |
Hb_001758_030 |
0.3021694002 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 8 |
Hb_003717_020 |
0.3166461232 |
- |
- |
- |
| 9 |
Hb_065395_020 |
0.3257109906 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 10 |
Hb_176819_010 |
0.3347894189 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 11 |
Hb_132317_010 |
0.3371864881 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 12 |
Hb_001021_050 |
0.3386553191 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 13 |
Hb_005241_020 |
0.3446834224 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 14 |
Hb_024468_040 |
0.3563568755 |
- |
- |
- |
| 15 |
Hb_003992_200 |
0.3591133056 |
- |
- |
- |
| 16 |
Hb_006040_040 |
0.3616075093 |
- |
- |
Putative mitochondrial protein [Glycine soja] |
| 17 |
Hb_027204_010 |
0.3620719306 |
- |
- |
- |
| 18 |
Hb_065661_010 |
0.3700173639 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 19 |
Hb_153142_010 |
0.3704723482 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 20 |
Hb_000140_430 |
0.3712452603 |
- |
- |
- |