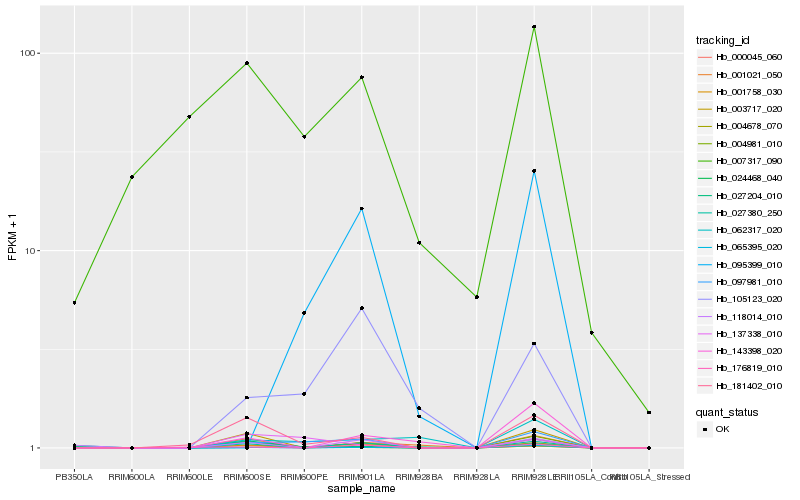
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001758_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 2 |
Hb_097981_010 |
0.1337335142 |
- |
- |
- |
| 3 |
Hb_004678_070 |
0.1969765516 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 4 |
Hb_004981_010 |
0.2292906618 |
- |
- |
- |
| 5 |
Hb_181402_010 |
0.2558420676 |
- |
- |
PREDICTED: uncharacterized protein LOC104905886 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_105123_020 |
0.2813306785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003717_020 |
0.2854181906 |
- |
- |
- |
| 8 |
Hb_065395_020 |
0.2939421745 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 9 |
Hb_118014_010 |
0.3021694002 |
- |
- |
PREDICTED: uncharacterized protein LOC104214283 [Nicotiana sylvestris] |
| 10 |
Hb_001021_050 |
0.3174017931 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 11 |
Hb_000045_060 |
0.3181261911 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_176819_010 |
0.3256601134 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 13 |
Hb_062317_020 |
0.3433662921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_024468_040 |
0.3460132503 |
- |
- |
- |
| 15 |
Hb_007317_090 |
0.349648868 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 16 |
Hb_143398_020 |
0.3519786288 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 17 |
Hb_095399_010 |
0.3530340555 |
- |
- |
cytochrome b/f complex subunit IV [Sphagnum fimbriatum] |
| 18 |
Hb_137338_010 |
0.3537943947 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_027380_250 |
0.3542240293 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 20 |
Hb_027204_010 |
0.3568242619 |
- |
- |
- |