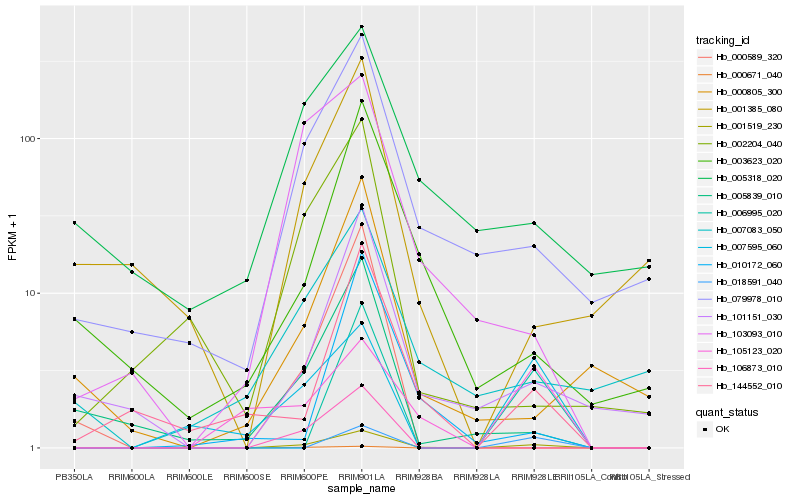
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001519_230 |
0.0 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 2 |
Hb_079978_010 |
0.2521883932 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 3 |
Hb_101151_030 |
0.2749121078 |
- |
- |
- |
| 4 |
Hb_144552_010 |
0.275924765 |
- |
- |
CC-NBS-LRR protein, putative [Theobroma cacao] |
| 5 |
Hb_007595_060 |
0.2766233887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_106873_010 |
0.2773541293 |
- |
- |
- |
| 7 |
Hb_006995_020 |
0.2884042557 |
- |
- |
hypothetical protein glysoja_017339 [Glycine soja] |
| 8 |
Hb_005839_010 |
0.2926076444 |
- |
- |
30S ribosomal protein s13 [Camellia sinensis] |
| 9 |
Hb_103093_010 |
0.2935434705 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 10 |
Hb_002204_040 |
0.2960250561 |
- |
- |
hypothetical protein B456_008G181200 [Gossypium raimondii] |
| 11 |
Hb_000589_320 |
0.2999308326 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_003623_020 |
0.3009007703 |
- |
- |
- |
| 13 |
Hb_000671_040 |
0.3052656018 |
- |
- |
- |
| 14 |
Hb_007083_050 |
0.3060949187 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 15 |
Hb_000805_300 |
0.3099712773 |
- |
- |
- |
| 16 |
Hb_105123_020 |
0.3122539145 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010172_060 |
0.314384166 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 18 |
Hb_005318_020 |
0.3143940991 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 19 |
Hb_018591_040 |
0.3174447872 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 20 |
Hb_001385_080 |
0.3178906595 |
- |
- |
- |