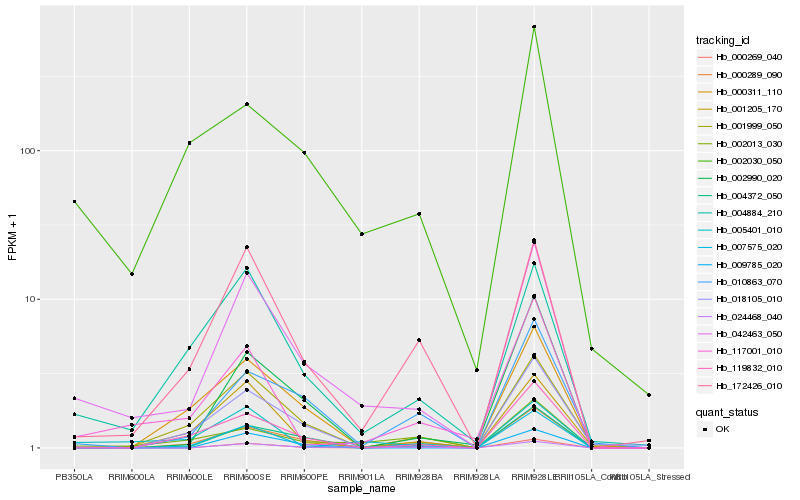
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042463_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004884_210 |
0.1856421787 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 3 |
Hb_024468_040 |
0.1865042817 |
- |
- |
- |
| 4 |
Hb_172426_010 |
0.1929569807 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 5 |
Hb_004372_050 |
0.2002342567 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 6 |
Hb_001999_050 |
0.2032907106 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 7 |
Hb_002030_050 |
0.2049169913 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 8 |
Hb_000311_110 |
0.2081108144 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_007575_020 |
0.2108629371 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_117001_010 |
0.2108633008 |
- |
- |
- |
| 11 |
Hb_010863_070 |
0.2116631119 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 12 |
Hb_001205_170 |
0.2131938571 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002990_020 |
0.2168719943 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 14 |
Hb_000289_090 |
0.2178160233 |
- |
- |
- |
| 15 |
Hb_002013_030 |
0.2197080477 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 16 |
Hb_119832_010 |
0.2207333879 |
- |
- |
- |
| 17 |
Hb_005401_010 |
0.2243540327 |
- |
- |
- |
| 18 |
Hb_018105_010 |
0.227850548 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 19 |
Hb_000269_040 |
0.2307334991 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_009785_020 |
0.2319630191 |
- |
- |
PREDICTED: nucleolin [Populus euphratica] |