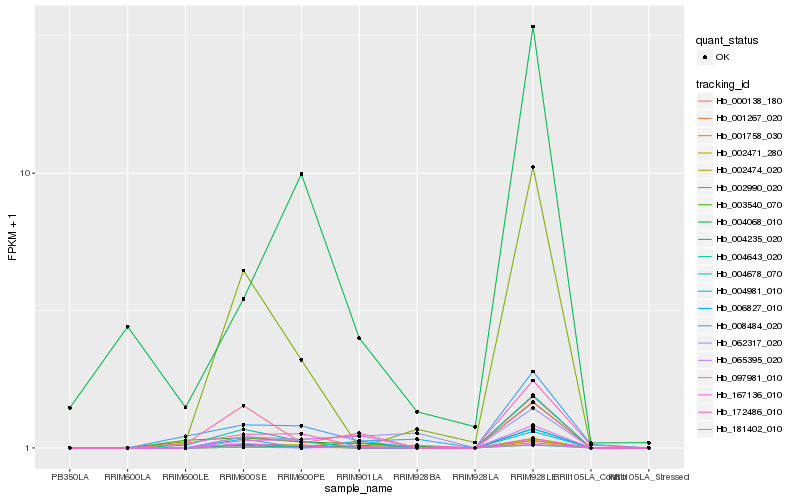
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004678_070 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 2 |
Hb_097981_010 |
0.1405410607 |
- |
- |
- |
| 3 |
Hb_001758_030 |
0.1969765516 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 4 |
Hb_065395_020 |
0.2480414458 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_003540_070 |
0.2768981879 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 6 |
Hb_004981_010 |
0.2791391581 |
- |
- |
- |
| 7 |
Hb_002471_280 |
0.2792979377 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 8 |
Hb_181402_010 |
0.2810370754 |
- |
- |
PREDICTED: uncharacterized protein LOC104905886 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_000138_180 |
0.2827070789 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 10 |
Hb_167136_010 |
0.2829815788 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_001267_020 |
0.2856377562 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 12 |
Hb_004068_010 |
0.2921379511 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 13 |
Hb_006827_010 |
0.2962572994 |
- |
- |
retrotransposon protein, putative, unclassified, expressed [Oryza sativa Japonica Group] |
| 14 |
Hb_062317_020 |
0.2962932877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008484_020 |
0.297215933 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 16 |
Hb_002990_020 |
0.2978311217 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 17 |
Hb_002474_020 |
0.3026617197 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104585652 [Nelumbo nucifera] |
| 18 |
Hb_004643_020 |
0.3034574943 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 19 |
Hb_004235_020 |
0.3037267823 |
- |
- |
PREDICTED: uncharacterized protein LOC103322555 [Prunus mume] |
| 20 |
Hb_172486_010 |
0.3037618605 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |