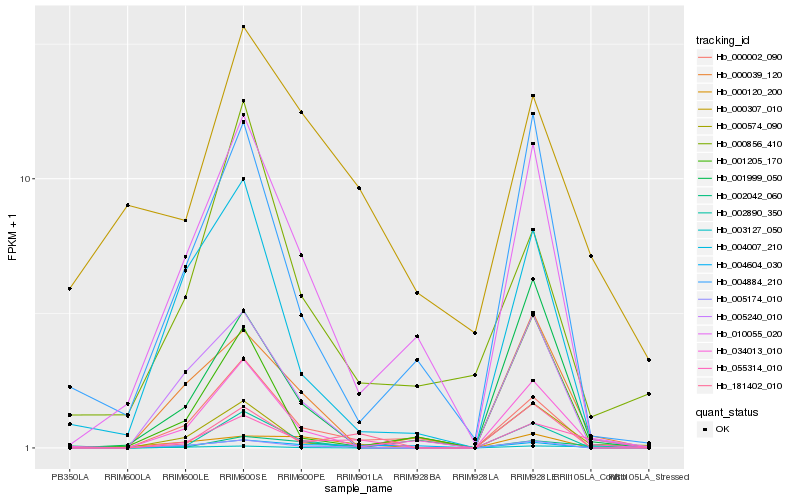
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055314_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_181402_010 |
0.2537748714 |
- |
- |
PREDICTED: uncharacterized protein LOC104905886 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_003127_050 |
0.2658044078 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 4 |
Hb_034013_010 |
0.2799021838 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 5 |
Hb_000002_090 |
0.2928460999 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000039_120 |
0.3042185291 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 7 |
Hb_010055_020 |
0.3062091263 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 8 |
Hb_005174_010 |
0.3075532172 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000574_090 |
0.3137776834 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 10 |
Hb_001999_050 |
0.3189230458 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 11 |
Hb_000307_010 |
0.3206649229 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 12 |
Hb_004007_210 |
0.3226680875 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 13 |
Hb_004604_030 |
0.3236954317 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_002042_060 |
0.3279366969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001205_170 |
0.3291827358 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004884_210 |
0.3295369349 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 17 |
Hb_000120_200 |
0.3307445828 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 18 |
Hb_000856_410 |
0.3313860899 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_005240_010 |
0.3319088407 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 20 |
Hb_002890_350 |
0.3324187545 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |