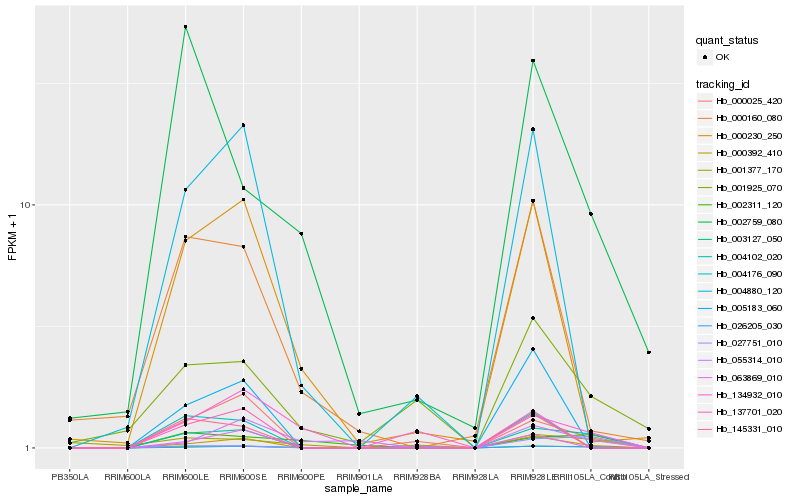
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004102_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_003127_050 |
0.1939298145 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_000392_410 |
0.2457759763 |
- |
- |
PREDICTED: uncharacterized protein LOC104882159 [Vitis vinifera] |
| 4 |
Hb_000025_420 |
0.2684293449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002311_120 |
0.318999012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_026205_030 |
0.327186675 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_137701_020 |
0.339462554 |
- |
- |
putative protein kinase [Oryza sativa Japonica Group] |
| 8 |
Hb_004176_090 |
0.3407260299 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 9 |
Hb_001925_070 |
0.3430167713 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 10 |
Hb_145331_010 |
0.3473095792 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0016s07690g [Populus trichocarpa] |
| 11 |
Hb_000160_080 |
0.3582247174 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM2b-like [Jatropha curcas] |
| 12 |
Hb_005183_060 |
0.358286267 |
- |
- |
hypothetical protein JCGZ_04271 [Jatropha curcas] |
| 13 |
Hb_004880_120 |
0.3600057238 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 14 |
Hb_063869_010 |
0.3601155909 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 15 |
Hb_000230_250 |
0.3622821454 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002759_080 |
0.3630480097 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 17 |
Hb_027751_010 |
0.3673915959 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_055314_010 |
0.371808767 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_001377_170 |
0.374742557 |
- |
- |
hypothetical protein JCGZ_14264 [Jatropha curcas] |
| 20 |
Hb_134932_010 |
0.3756791727 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |