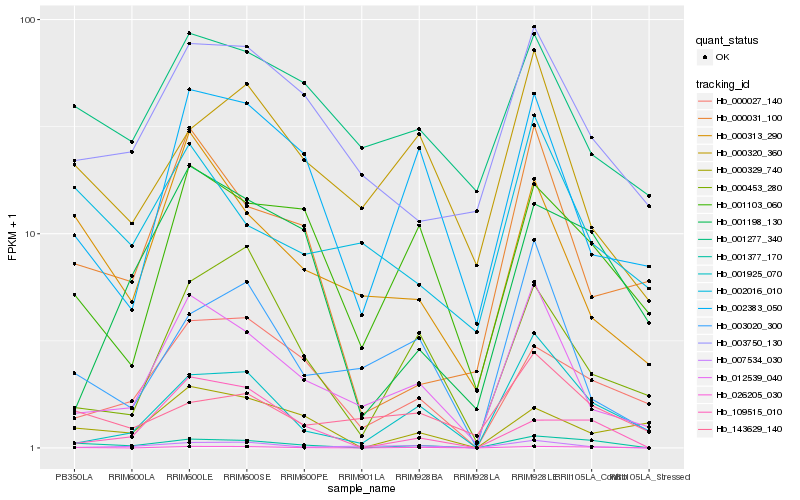
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_170 |
0.0 |
- |
- |
hypothetical protein JCGZ_14264 [Jatropha curcas] |
| 2 |
Hb_026205_030 |
0.184693578 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001925_070 |
0.2692003114 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 4 |
Hb_143629_140 |
0.2774611853 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_001103_060 |
0.2789205609 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 6 |
Hb_000027_140 |
0.2825698753 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 7 |
Hb_003750_130 |
0.2839899138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012539_040 |
0.2856962607 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 9 |
Hb_109515_010 |
0.2900583521 |
- |
- |
- |
| 10 |
Hb_000313_290 |
0.2929315376 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_002016_010 |
0.294006138 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001198_130 |
0.2944546681 |
- |
- |
PREDICTED: potassium channel AKT2/3 [Jatropha curcas] |
| 13 |
Hb_000031_100 |
0.2964107936 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 14 |
Hb_000320_360 |
0.2985259755 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000329_740 |
0.3013973876 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 16 |
Hb_001277_340 |
0.3015571947 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 17 |
Hb_003020_300 |
0.3019626016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007534_030 |
0.3033026563 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 19 |
Hb_002383_050 |
0.3038581692 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 20 |
Hb_000453_280 |
0.304092073 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |