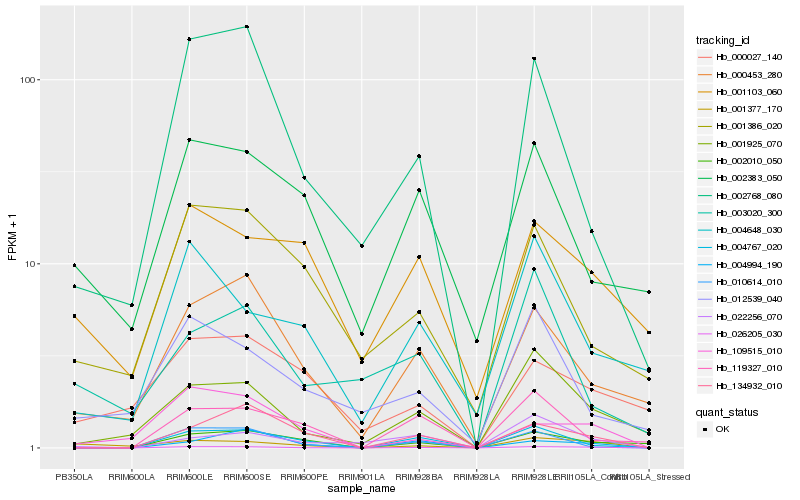
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026205_030 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_001377_170 |
0.184693578 |
- |
- |
hypothetical protein JCGZ_14264 [Jatropha curcas] |
| 3 |
Hb_000453_280 |
0.2484733695 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 4 |
Hb_001925_070 |
0.2553484687 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 5 |
Hb_004767_020 |
0.2636619109 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_001103_060 |
0.2662002894 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 7 |
Hb_134932_010 |
0.2697664264 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |
| 8 |
Hb_109515_010 |
0.2711494613 |
- |
- |
- |
| 9 |
Hb_002768_080 |
0.271570595 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_002383_050 |
0.2739734579 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 11 |
Hb_002010_050 |
0.2827301278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_022256_070 |
0.2835917769 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 13 |
Hb_001386_020 |
0.2837352592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004648_030 |
0.2889510817 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 15 |
Hb_012539_040 |
0.2906555745 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 16 |
Hb_004994_190 |
0.2913156198 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Jatropha curcas] |
| 17 |
Hb_010614_010 |
0.2949943727 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 18 |
Hb_000027_140 |
0.2975712938 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 19 |
Hb_003020_300 |
0.2984295856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_119327_010 |
0.3006032824 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |