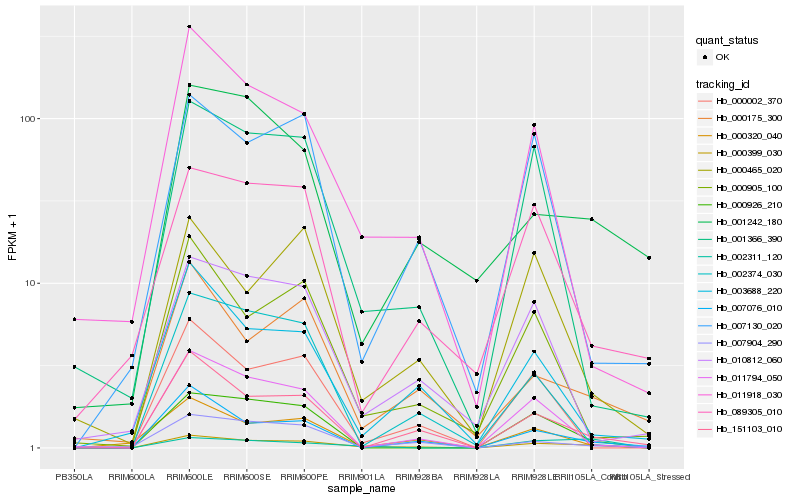
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000399_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 2 |
Hb_151103_010 |
0.1906966661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007076_010 |
0.1994003804 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_011794_050 |
0.2018107377 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000320_040 |
0.2060657003 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000926_210 |
0.2063373886 |
- |
- |
LIM and calponin domains-containing protein 1, putative [Theobroma cacao] |
| 7 |
Hb_001366_390 |
0.2095146266 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 8 |
Hb_011918_030 |
0.215140871 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 9 |
Hb_000175_300 |
0.2176541363 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 10 |
Hb_003688_220 |
0.2224922524 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_000905_100 |
0.2226425008 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_089305_010 |
0.2248219491 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 13 |
Hb_000465_020 |
0.2252352412 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 14 |
Hb_007904_290 |
0.2254456759 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 15 |
Hb_010812_060 |
0.2259809378 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 16 |
Hb_000002_370 |
0.2260392255 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 17 |
Hb_007130_020 |
0.2268754161 |
- |
- |
- |
| 18 |
Hb_001242_180 |
0.2293196291 |
- |
- |
choline/ethanolamine kinase, putative [Ricinus communis] |
| 19 |
Hb_002374_030 |
0.2299026664 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002311_120 |
0.2318591923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |