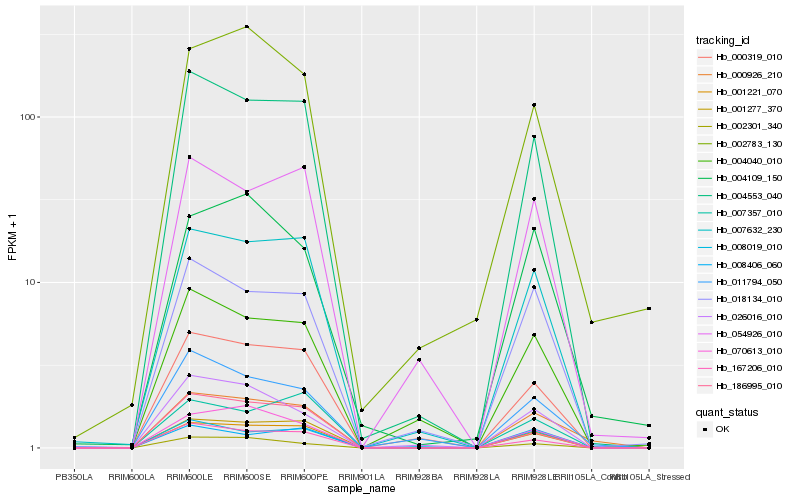
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_210 |
0.0 |
- |
- |
LIM and calponin domains-containing protein 1, putative [Theobroma cacao] |
| 2 |
Hb_001221_070 |
0.1243300772 |
- |
- |
PREDICTED: calcium-binding protein CML38-like [Jatropha curcas] |
| 3 |
Hb_004553_040 |
0.1307642136 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 4 |
Hb_001277_370 |
0.1364690672 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 5 |
Hb_007632_230 |
0.1389401317 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 6 |
Hb_167206_010 |
0.1480924468 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 7 |
Hb_004109_150 |
0.1506522733 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 8 |
Hb_000319_010 |
0.1544785664 |
- |
- |
hypothetical protein POPTR_0016s03600g [Populus trichocarpa] |
| 9 |
Hb_011794_050 |
0.1549908183 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_018134_010 |
0.1550104799 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 11 |
Hb_070613_010 |
0.1574422389 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_054926_010 |
0.1593177263 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 13 |
Hb_002783_130 |
0.1600057786 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_004040_010 |
0.1616196257 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 15 |
Hb_002301_340 |
0.1641580141 |
transcription factor |
TF Family: B3 |
leafy cotyledon 2 [Ricinus communis] |
| 16 |
Hb_008019_010 |
0.1680617994 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 17 |
Hb_026016_010 |
0.1717038344 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_008406_060 |
0.1730369123 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 19 |
Hb_007357_010 |
0.1749312311 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_186995_010 |
0.1753088396 |
- |
- |
hypothetical protein POPTR_0012s01755g [Populus trichocarpa] |