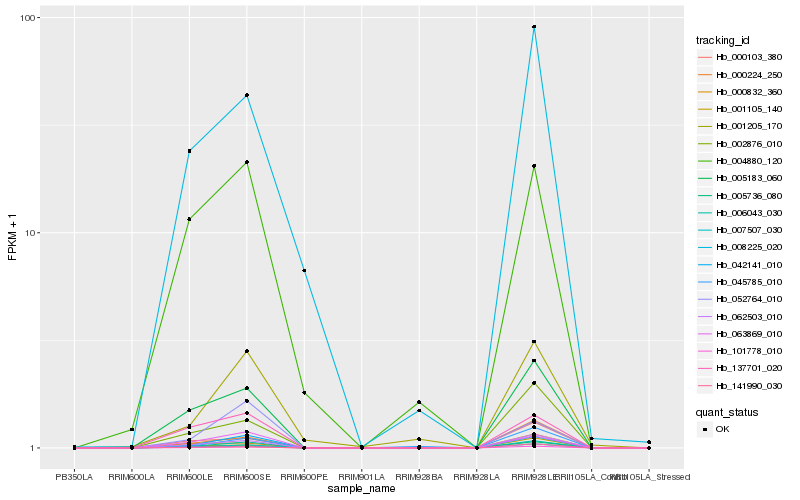
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007507_030 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_101778_010 |
0.0294450179 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_062503_010 |
0.0765900583 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_042141_010 |
0.0855031317 |
- |
- |
- |
| 5 |
Hb_005183_060 |
0.0976386548 |
- |
- |
hypothetical protein JCGZ_04271 [Jatropha curcas] |
| 6 |
Hb_000224_250 |
0.1402297614 |
- |
- |
hypothetical protein AMTR_s05400p00003230 [Amborella trichopoda] |
| 7 |
Hb_063869_010 |
0.1410531531 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 8 |
Hb_002876_010 |
0.147085827 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_137701_020 |
0.154860061 |
- |
- |
putative protein kinase [Oryza sativa Japonica Group] |
| 10 |
Hb_000103_380 |
0.1624358033 |
- |
- |
PREDICTED: uncharacterized protein LOC104596624 [Nelumbo nucifera] |
| 11 |
Hb_008225_020 |
0.1759877079 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 12 |
Hb_000832_360 |
0.179258794 |
- |
- |
- |
| 13 |
Hb_001205_170 |
0.1799641282 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001105_140 |
0.1861413752 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_004880_120 |
0.1907748539 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 16 |
Hb_005736_080 |
0.1911180683 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 17 |
Hb_141990_030 |
0.193782434 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 18 |
Hb_052764_010 |
0.1951205517 |
- |
- |
hypothetical protein POPTR_0011s04320g [Populus trichocarpa] |
| 19 |
Hb_045785_010 |
0.1975856984 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 20 |
Hb_006043_030 |
0.1999744897 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |