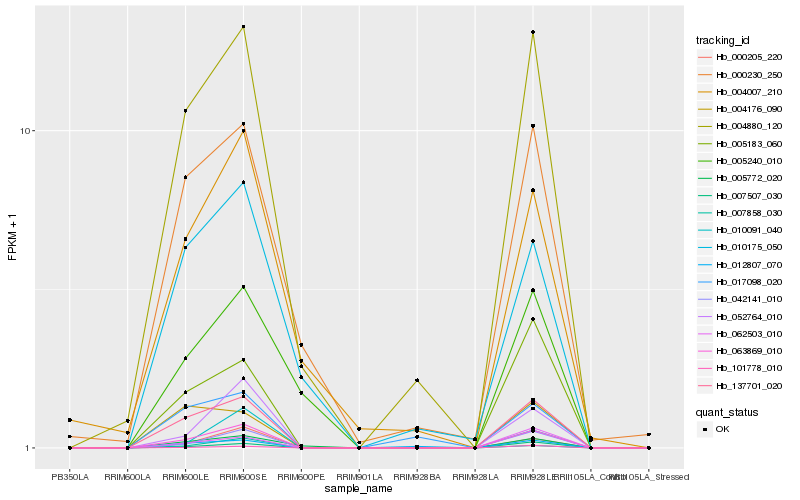
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063869_010 |
0.0 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 2 |
Hb_137701_020 |
0.0806882042 |
- |
- |
putative protein kinase [Oryza sativa Japonica Group] |
| 3 |
Hb_042141_010 |
0.0977224697 |
- |
- |
- |
| 4 |
Hb_000205_220 |
0.1145218917 |
- |
- |
PREDICTED: uncharacterized protein LOC105647760 [Jatropha curcas] |
| 5 |
Hb_052764_010 |
0.1206474764 |
- |
- |
hypothetical protein POPTR_0011s04320g [Populus trichocarpa] |
| 6 |
Hb_007507_030 |
0.1410531531 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_004880_120 |
0.1481005688 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 8 |
Hb_101778_010 |
0.163536065 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_005183_060 |
0.1662305463 |
- |
- |
hypothetical protein JCGZ_04271 [Jatropha curcas] |
| 10 |
Hb_010091_040 |
0.1748841843 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 11 |
Hb_062503_010 |
0.1871881665 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_007858_030 |
0.1925603532 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 13 |
Hb_005772_020 |
0.1943216193 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 14 |
Hb_000230_250 |
0.207924981 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_012807_070 |
0.209090326 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 16 |
Hb_004176_090 |
0.2098945728 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 17 |
Hb_004007_210 |
0.2103097127 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 18 |
Hb_010175_050 |
0.2128716352 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 19 |
Hb_005240_010 |
0.2168023707 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 20 |
Hb_017098_020 |
0.2183690306 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |