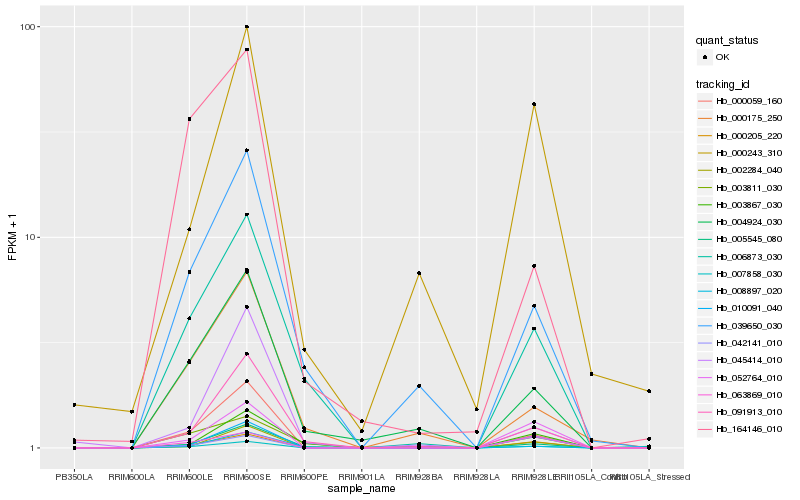
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007858_030 |
0.0 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 2 |
Hb_000205_220 |
0.0977270221 |
- |
- |
PREDICTED: uncharacterized protein LOC105647760 [Jatropha curcas] |
| 3 |
Hb_000059_160 |
0.1113657457 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 4 |
Hb_010091_040 |
0.1357391238 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 5 |
Hb_052764_010 |
0.1421453161 |
- |
- |
hypothetical protein POPTR_0011s04320g [Populus trichocarpa] |
| 6 |
Hb_002284_040 |
0.1469274292 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_091913_010 |
0.1793331468 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 8 |
Hb_006873_030 |
0.1872254845 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 9 |
Hb_063869_010 |
0.1925603532 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 10 |
Hb_004924_030 |
0.1937972221 |
- |
- |
- |
| 11 |
Hb_008897_020 |
0.2015821916 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 12 |
Hb_039650_030 |
0.2019853906 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 13 |
Hb_003867_030 |
0.2238177238 |
- |
- |
- |
| 14 |
Hb_000175_250 |
0.2238730292 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 15 |
Hb_000243_310 |
0.2325665172 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 16 |
Hb_164146_010 |
0.2352191028 |
- |
- |
- |
| 17 |
Hb_045414_010 |
0.2374314443 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_003811_030 |
0.2392043434 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 19 |
Hb_042141_010 |
0.2392788484 |
- |
- |
- |
| 20 |
Hb_005545_080 |
0.2398009089 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |