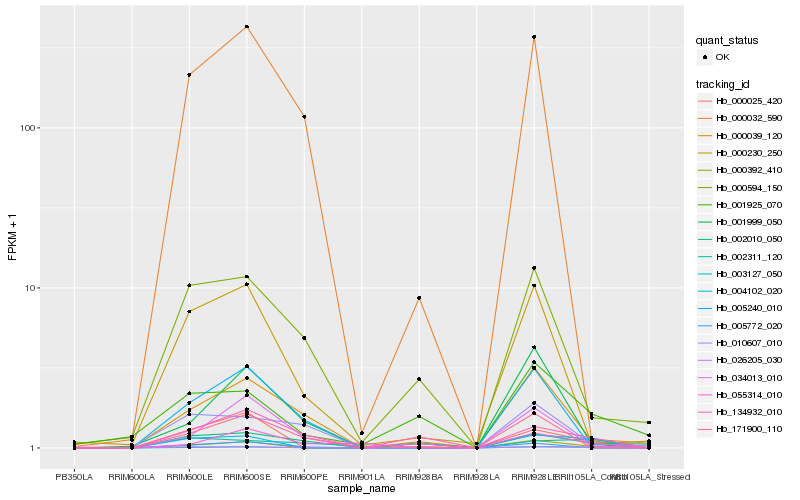
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003127_050 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_004102_020 |
0.1939298145 |
- |
- |
- |
| 3 |
Hb_055314_010 |
0.2658044078 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_002311_120 |
0.2679462576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000392_410 |
0.2722413583 |
- |
- |
PREDICTED: uncharacterized protein LOC104882159 [Vitis vinifera] |
| 6 |
Hb_000039_120 |
0.27486579 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 7 |
Hb_134932_010 |
0.2891728273 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1, partial [Populus euphratica] |
| 8 |
Hb_010607_010 |
0.3104025548 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 9 |
Hb_026205_030 |
0.3124897372 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_000025_420 |
0.314852185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002010_050 |
0.3180069427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005240_010 |
0.3205471262 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 13 |
Hb_001999_050 |
0.3235539716 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 14 |
Hb_001925_070 |
0.3255498459 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 15 |
Hb_034013_010 |
0.3256662733 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 16 |
Hb_005772_020 |
0.3271592791 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 17 |
Hb_171900_110 |
0.3289412511 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 18 |
Hb_000032_590 |
0.330425294 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 19 |
Hb_000230_250 |
0.331216746 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000594_150 |
0.3353393722 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |