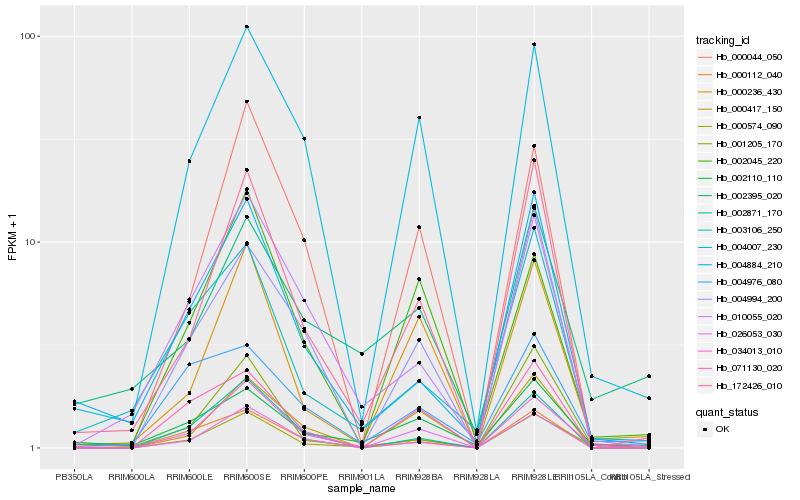
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 2 |
Hb_172426_010 |
0.1607904415 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 3 |
Hb_000236_430 |
0.1622195968 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 4 |
Hb_034013_010 |
0.1711758551 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 5 |
Hb_001205_170 |
0.1833996063 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004007_230 |
0.1849202254 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002110_110 |
0.1862696285 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 8 |
Hb_002871_170 |
0.189106192 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 9 |
Hb_004994_200 |
0.1907206416 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 10 |
Hb_004884_210 |
0.1943631188 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 11 |
Hb_003106_250 |
0.1969978616 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 12 |
Hb_000112_040 |
0.1994755158 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_026053_030 |
0.2035686019 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 14 |
Hb_010055_020 |
0.2055027065 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 15 |
Hb_071130_020 |
0.2056862451 |
- |
- |
- |
| 16 |
Hb_002395_020 |
0.2082124154 |
- |
- |
Protein CREG1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000044_050 |
0.2083153591 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000417_150 |
0.2090442153 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 19 |
Hb_004976_080 |
0.2095147128 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 20 |
Hb_002045_220 |
0.2103030298 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |