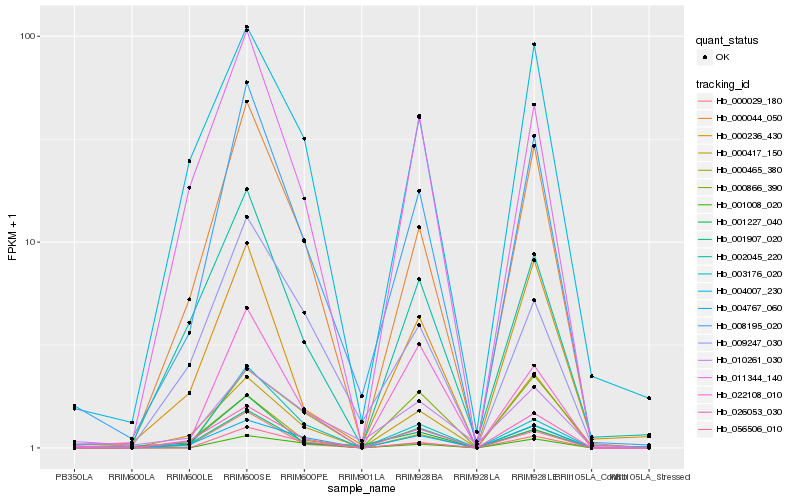
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008195_020 |
0.0 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 2 |
Hb_000044_050 |
0.0963169181 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_026053_030 |
0.1321103564 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 4 |
Hb_001907_020 |
0.1393985539 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 5 |
Hb_011344_140 |
0.1487933167 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 6 |
Hb_000417_150 |
0.1489962216 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 7 |
Hb_000236_430 |
0.150912296 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 8 |
Hb_000465_380 |
0.153711463 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002045_220 |
0.1537892519 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001227_040 |
0.153904251 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 11 |
Hb_004007_230 |
0.1590002254 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009247_030 |
0.1626464572 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 13 |
Hb_022108_010 |
0.1645734975 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Glycine soja] |
| 14 |
Hb_056506_010 |
0.1661119097 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 15 |
Hb_004767_060 |
0.1683627411 |
- |
- |
PREDICTED: uncharacterized protein LOC105761928 [Gossypium raimondii] |
| 16 |
Hb_010261_030 |
0.1710074787 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 17 |
Hb_003176_020 |
0.1720066763 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 18 |
Hb_000029_180 |
0.1733085099 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 19 |
Hb_000866_390 |
0.1740964886 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 20 |
Hb_001008_020 |
0.1752835027 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |